This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
High-resolution roadmap charted for regeneration of pancreatic beta-cells
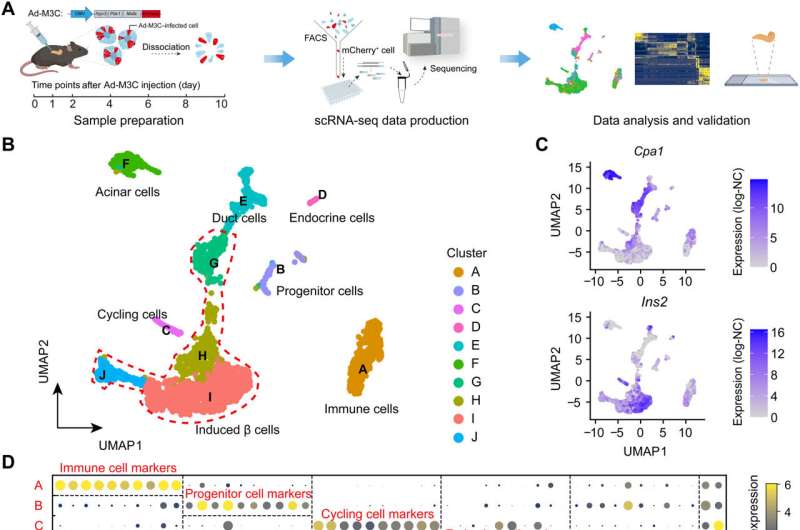
A research group led by Prof. Shao Zhen from the Shanghai Institute of Nutrition and Health (SINH) of the Chinese Academy of Sciences (CAS) and the collaborators charted a high-resolution roadmap for the process of pancreatic β cell regeneration by in vivo transdifferentiation from adult acinar cells using single-cell RNA sequencing (scRNA-seq) technology. The study was published in Science Advances.
Adult mammals have limited capacity to regenerate functional cells. In vivo transdifferentiation heralds the possibility of regeneration by lineage reprogramming from other fully differentiated cells. Taking pancreatic β cell regeneration as a paradigm, it has been reported that in vivo transdifferentiation from adult mouse pancreatic acinar cells to induced β cells has been achieved which is induced by Mafa, Pdx1, and Ngn3 (M3 factor).
Regenerated β cells in this way could produce and secrete insulin without causing rejection in mice, which holds great promise for curing diabetes.
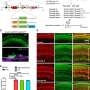
Acinar cells are the most abundant cell type in pancreas, accounting for up to 99%, which can serve as an ideal source for β cell regeneration. Pancreatic microenvironment can also promote the maturation of regenerated β cells and the formation of islet-like structures. However, the current understanding of the in vivo transdifferentiation process from acinar cells to β cells is still superficial. How the cell fate changes and what factors play key roles during this process need further research and interpretation.
The single-cell transcriptomic study of in vivo transdifferentiation from adult mouse acinar cells to induced β cells was carried out by Prof. Shao's group and Prof. Li Weida's group from Tongji University.
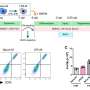
The researchers revealed that the constructed cell trajectory of the transdifferentiation process could be divided into two stages, stage 1 which is single line, and stage 2 which is bifurcated. The two lineages of stage 2 correspond to different cell fates. One of them leads to successfully reprogrammed β cells and the other one leads to the dead-end.
Successive study of these two stages revealed that p53 plays a role of reprogramming barrier by regulating cell cycle arrest in stage 1, and high expression of Dnmt3a leads cells to the fate of reprogramming failure in stage 2. Moreover, deletion of p53 or reduction of Dnmt3a expression increased the efficiency of β cell regeneration.
This study deciphered a high-resolution roadmap for regeneration of β cells by in vivo transdifferentiation from adult acinar cells, and identified a series of important regulatory factors, which provides a detailed molecular blueprint to facilitate in vivo regeneration in mammals.
More information: Gang Liu et al, Charting a high-resolution roadmap for regeneration of pancreatic β cells by in vivo transdifferentiation from adult acinar cells, Science Advances (2023). DOI: 10.1126/sciadv.adg2183
Journal information: Science Advances
Provided by Chinese Academy of Sciences