This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Using AI to create better, more potent medicines
While it can take years for the pharmaceutical industry to create medicines capable of treating or curing human disease, a new study suggests that using generative artificial intelligence could vastly accelerate the drug-development process.
Today, most drug discovery is carried out by human chemists who rely on their knowledge and experience to select and synthesize the right molecules needed to become the safe and efficient medicines we depend on. To identify the synthesis paths, scientists often employ a technique called retrosynthesis—a method for creating potential drugs by working backward from the wanted molecules and searching for chemical reactions to make them.
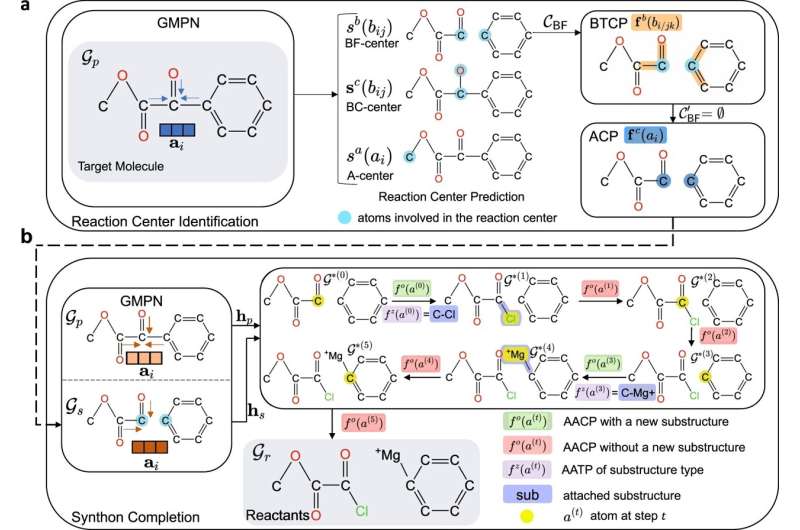
Yet because sifting through millions of potential chemical reactions can be an extremely challenging and time-consuming endeavor, researchers at The Ohio State University have created an AI framework called G2Retro to automatically generate reactions for any given molecule. The new study showed that compared to current manual-planning methods, the framework was able to cover an enormous range of possible chemical reactions as well as accurately and quickly discern which reactions might work best to create a given drug molecule.
"Using AI for things critical to saving human lives, such as medicine, is what we really want to focus on," said Xia Ning, lead author of the study and an associate professor of computer science and engineering at Ohio State. "Our aim was to use AI to accelerate the drug design process, and we found that it not only saves researchers time and money but provides drug candidates that may have much better properties than any molecules that exist in nature."
This study builds on previous research of Ning's where her team developed a method named Modof that was able to generate molecule structures that exhibited desired properties better than any existing molecules. "Now the question becomes how to make such generated molecules, and that is where this new study shines," said Ning, also an associate professor of biomedical informatics in the College of Medicine.
The study was published today in the journal Communications Chemistry.
Ning's team trained G2Retro on a dataset that contains 40,000 chemical reactions collected between 1976 and 2016. The framework "learns" from graph-based representations of given molecules, and uses deep neural networks to generate possible reactant structures that could be used to synthesize them. Its generative power is so impressive that, according to Ning, once given a molecule, G2Retro could come up with hundreds of new reaction predictions in only a few minutes.
"Our generative AI method G2Retro is able to supply multiple different synthesis routes and options, as well as a way to rank different options for each molecule," said Ning. "This is not going to replace current lab-based experiments, but it will offer more and better drug options so experiments can be prioritized and focused much faster."
To further test the AI's effectiveness, Ning's team conducted a case study to see if G2Retro could accurately predict four newly released drugs already in circulation: Mitapivat, a medication used to treat hemolytic anemia; Tapinarof, which is used to treat various skin diseases; Mavacamten, a drug to treat systemic heart failure; and Oteseconazole, used to treat fungal infections in females. G2Retro was able to correctly generate exactly the same patented synthesis routes for these medicines, and provided alternative synthesis routes that are also feasible and synthetically useful, Ning said.
Having such a dynamic and effective device at scientists' disposal could enable the industry to manufacture stronger drugs at a quicker pace—but despite the edge AI might give scientists inside the lab, Ning emphasizes the medicines G2Retro or any generative AI creates still need to be validated—a process that involves the created molecules being tested in animal models and later in human trials.
"We are very excited about generative AI for medicine, and we are dedicated to using AI responsibly to improve human health," said Ning.
More information: Ziqi Chen et al, G2Retro as a two-step graph generative models for retrosynthesis prediction, Communications Chemistry (2023). DOI: 10.1038/s42004-023-00897-3
Provided by The Ohio State University