This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Study reveals new mechanism for DNA folding
A hitherto unknown mechanism for DNA folding is described in a study in Nature published by researchers from Karolinska Institutet and the Max Planck Institute for Biophysics. Their findings provide new insights into chromosomal processes that are vital to both normal development and to prevent disease.
The DNA in our cells is organized into chromosomes, which are highly dynamic structures that are altered when genes are transcribed, when DNA damage is repaired or when chromosomes are compacted in preparation for cell division. These processes are affected by so called SMC protein complexes (SMC, Structural Maintenance of Chromosomes), which by mediating chromosomal interactions ensure correct spatial organization of the genome.
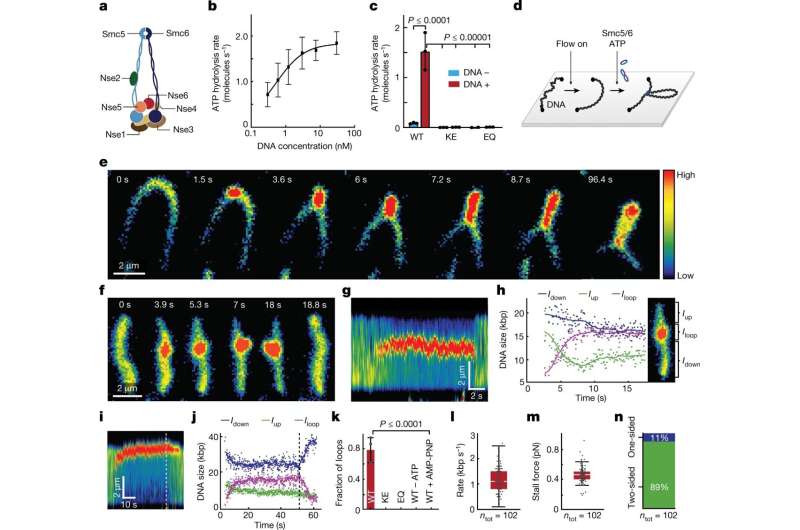
In humans and other eukaryotes, i.e., organisms whose cells contain a nucleus, there are three such protein complexes. Scientists have already revealed the mechanism of function for two of them. In the present study, the researchers investigated the third, the Smc5/6 complex, the function of which has remained mostly unknown.
"These results reveal the Smc5/6 complex as a new regulator of DNA folding, which can tell us more about how chromosomes are organized," says Camilla Björkegren, professor at the Department of Cell and Molecular Biology at Karolinska Institutet, who led the study together with Eugene Kim, research group leader at the Max Planck Institute for Biophysics in Frankfurt am Main.
"The discovery is also medically relevant, since DNA folding is important for normal chromosome function and for avoiding chromosomal alterations that could lead to disease."
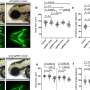
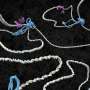
The researchers have purified the Smc5/6 complex from yeast and, using high-resolution microscopy of individual molecules, have studied how it binds and affects individual DNA molecules. The principles of chromosomal organization are believed to be generally identical in yeast and humans, which are both eukaryotic organisms. For their experiments, the researchers both protein complex and DNA were labeled with differently colored fluorescing molecules to make them traceable through a microscope.
Their results show that the Smc5/6 complex operates by extruding an increasingly larger DNA loop, a property it shares with the other known eukaryotic SMC complexes.
The researchers have also examined how the process is regulated and found, among other things, that two Smc5/6 complexes are needed to form a loop, while single protein complexes only translocates along the DNA molecule.
Previous research indicates that Smc5/6 inhibits certain viruses and suggests that it also protects against certain types of cancer, and is important to normal fetal development. The KI researchers now want to study how these properties are related to the newly discovered mechanism.
"The next step of our research is to find out how the Smc5/6 complex's ability to make DNA loops affects their function in cells, which can increase our understanding of how Smc5/6 can function as a virus blocker, protect against cancer and contribute to fetal development," says Professor Björkegren.
More information: Eugene Kim, The Smc5/6 complex is a DNA loop extruding motor, Nature (2023). DOI: 10.1038/s41586-023-05963-3. www.nature.com/articles/s41586-023-05963-3
Journal information: Nature
Provided by Karolinska Institutet