This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
trusted source
proofread
Evolutionary trade-offs: Balancing genetic disease risks and pathogen protection
Certain genes can exist in different, functionally divergent variants in the individuals of a species. If they lead to clear differences, for example in a phenotype or susceptibility to disease, this is known in biology as a polymorphism. Typical examples are a number of genes whose variants are responsible for the expression of different blood groups. Interestingly, these polymorphic genes can be relevant to blood groups and diseases at the same time.
A research team from Kiel University and the Max Planck Institute for Evolutionary Biology (MPI-EB) led by Professor John Baines has now used mice as an example to investigate one such polymorphic gene, called B4galnt2, which in the individual animal can affect blood vessels and/or intestinal cells.
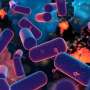
In this new study, the researchers from the Section of Evolutionary Medicine at Kiel University and MPI-EB were able to show that depending on the variant, the gene can cause not only a blood coagulation disorder, but also better immunity to bacterial infections. The researchers, who are active in the Collaborative Research Center (CRC) 1182 Origin and Function of Metaorganisms and the Cluster of Excellence Precision Medicine in Chronic Inflammation (PMI), succeeded in demonstrating the involvement of an individual variant of B4galnt2 in the defense against pathogens by identifying a previously unknown bacterium from the genus Morganella.
This group of microorganisms can cause problematic infections in humans, especially in hospitals and care facilities. In a so-called pathometagenomic analysis, the researchers showed that the prevalence of this bacterium relevant to inflammation is strongly restricted in mice with the B4galnt2 genotype affecting the blood vessels. They thus succeeded in elucidating an exemplary evolutionary trade-off between the risk for genetic disease and the evolutionary advantage of pathogen resistance that mice gain by retaining polymorphism at this gene. The researchers recently published their results in the scientific journal Gut Microbes.
Balancing evolutionary trade-offs
Over the course of evolution, certain genetic variants important for immune defense are favored under the selective pressure of infectious diseases, and the corresponding variants are therefore maintained. Retaining such genes, however, is often linked to so-called evolutionary trade-offs: Mice are known for example to have kept alternative forms (i.e. alleles) of the blood group-related B4galnt2 gene for almost three million years, despite it causing a bleeding time phenotype similar to von Willebrand disease in humans, which causes prolonged bleeding after injury.
"The maintenance of such a genetic variant must be associated with a strong selective advantage in other contexts, which were until now unknown," explains evolutionary biologist Baines. "Recent advances in the scientific understanding of the blood clotting system now suggest that genetic variation may also be involved in innate immunity and defense against pathogens, so we looked for a possible advantage of the B4galnt2 gene in this area," Baines continues.
Pathometagenomic analysis shows links between genetic variation and pathogen protection
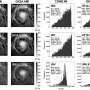
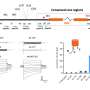
In order to investigate the role of variation at B4galnt2 in possible immunity to pathogens, the Kiel research team chose a new, so-called pathometagenomic approach: The researchers first examined the intestinal tissue of the animals for signs of inflammation. In the next step, they identified the microorganisms present in the animals' intestine via genome sequencing to detect correlations between microbiome composition and signs of inflammation.
"The overall composition of the microbiota does not seem to play a significant role at first. However, individual bacterial species were found to be disproportionately active in the presence of inflammation and particular genotypes at B4galnt2," says Baines.
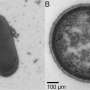
The researchers were able to narrow this observation to a previously unknown bacterial subspecies from the genus Morganella: Animals with the allele relevant for blood vessels and the associated risk for the blood clotting showed fewer signs of inflammation, with the bacterium almost being absent. In mice expressing B4galnt2 in the gastrointestinal tract, however, it is clearly detectable; the presence of inflammation here indicates its pathogenicity.
"While these animals do not carry the risk of the bleeding phenotype, the expression of B4galnt2 in the intestinal mucosa can be favored by pathogens. In the case of our analysis, it is Morganella that then leads to inflammation," says Baines.
Novel way to identify still unknown pathogens
In order to validate their findings in wild mice, the Kiel researchers collaborated with the group of Professor Guntram Grassl, a medical microbiologist at Hannover Medical School. The researchers then validated these findings derived from wild animals with infection experiments using mice in the laboratory that differed only according to the alleles present at the B4galnt2 gene. When these animals were inoculated with the bacterium, they showed the same signs of disease as the wild animals.
"This provides us with experimental evidence that the B4galnt2 gene plays an important role in susceptibility to bacterial infections in nature. This enabled us to validate that our novel pathometagenomic approach is in principle suitable for identifying yet unknown pathogens in wild animals, and thus for monitoring possible risks of such zoonotic pathogens for humans," says Grassl.
With the work now presented, the Kiel research team was also able to provide further evidence for the longstanding hypothesis on the evolutionary origins of blood group systems in general: "An important pioneer of evolutionary biology, the British geneticist J. B. S. Haldane, foresaw as early as the middle of the 20th century that blood groups and pathogen resistance may be related to one another. With research into blood group-related genes, which are a particularly common target of natural selection, numerous examples of this were described more recently," says Baines.
"However, the nature and extent of the evolutionary trade-offs involved are rarely explored in detail. With the help of our pathometagenomic analysis, we succeeded in linking pathogen resistance to a blood group-related gene, thus experimentally supporting Haldane's hypothesis."
More information: Marie Vallier et al, Pathometagenomics reveals susceptibility to intestinal infection by Morganella to be mediated by the blood group-related B4galnt2 gene in wild mice, Gut Microbes (2023). DOI: 10.1080/19490976.2022.2164448
Provided by Kiel University