This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
Elucidating enzyme gene expression in filamentous fungi for efficient biomass energy production
Filamentous fungi have long been a good friend of sake brewers, but they might soon also be a sidekick for environmentalists. Osaka Metropolitan University researchers have revealed the regulatory mechanisms of enzyme production in a filamentous fungus that allows for efficient degradation of plant biomass, an alternative energy resource to petroleum.
The research results were published in Applied Microbiology and Biotechnology on January 10, 2023.
Filamentous fungi (molds) are microorganisms with a long history of use in the fermentation of sake, soy sauce, cheese, and many other products. Such fermentation is a good example of the industrial use of filamentous fungi's ability to secrete various enzymes in large quantities.
Currently, plant biomass is attracting attention as an alternative to petroleum, which will eventually be depleted. Since hard plant cell walls are composed of various aromatics and polysaccharides, their degradation requires a large number of enzymes with diverse characteristics. Consequently, studies have been conducted to utilize filamentous fungi as a prominent source of enzymes for plant biomass degradation.
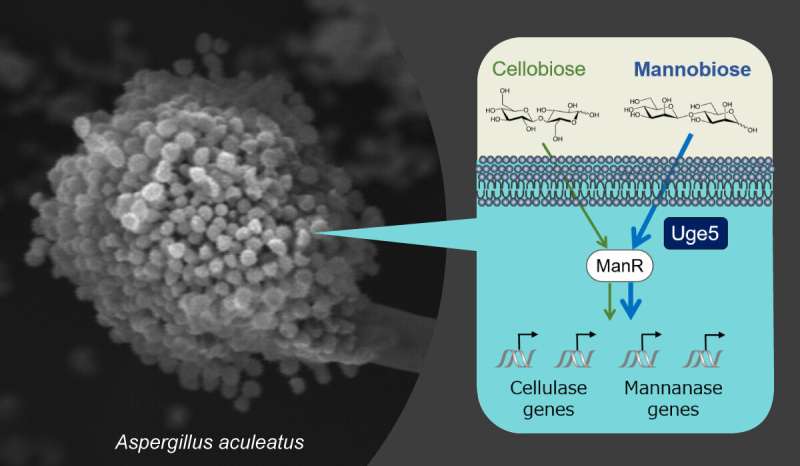
Delving into this field, a research team led by Associate Professor Shuji Tani, from the Graduate School of Agriculture at Osaka Metropolitan University, analyzed the regulatory mechanisms of carbohydrate-hydrolyzing enzyme production in the filamentous fungus Aspergillus aculeatus, which produces enzymes that have an excellent ability to degrade plant biomass.
Uridine diphosphate (UDP)-glucose 4-epimerase (Uge5) is well known as an enzyme involved in galactose metabolism. However, the team discovered that Uge5 also regulates the expression of degrading enzyme genes in A. aculeatus. This is the very first report of Uge5's roles in selective gene expression in response to different types of inducing sugars in filamentous fungi.
These findings address the current technology challenge in establishing a much-wanted comprehensive high production method for various enzymes in filamentous fungi.
Professor Tani explained, "We constructed and screened a library containing approximately 9,000 gene-disrupted strains of Aspergillus aculeatus, and identified Uge5 as a novel regulatory factor that regulates the production of carbohydrate-hydrolyzing enzymes. The discovery of this new function took us by surprise. We plan to continue our research to elucidate phenomena that existing knowledge cannot explain."
More information: A new function of a putative UDP-glucose 4-epimerase on the expression of glycoside hydrolase genes in Aspergillus aculeatus, Applied Microbiology and Biotechnology (2023). DOI: 10.1007/s00253-022-12337-8
Journal information: Applied Microbiology and Biotechnology
Provided by Osaka Metropolitan University