Universal multigene marker may revolutionize biodiversity research
An international team led by scientists from the Museum Koenig in Bonn (Leibniz Institute for the Analysis of Biodiversity Change) has for the first time successfully tested a universal set of genes for the systematic characterization of different animal species. Their work is published in Methods in Ecology and Evolution.
Many of the methods used to date for the molecular recording of biodiversity are still often flawed because they only use one genetic marker, which often does not correctly reflect species and genetic patterns at the genomic level.
Other analyses based on genomic data are not suitable for comprehensive biodiversity research because they are not universal enough and often not very sustainable because they are based on data that cannot be combined with the results of other studies.
The tested gene set, more precisely called "metazoan-level universal single-copy orthologs" (metazoan USCOs), comprises several hundred nuclear genes and can be used for all multicellular animals. The method clearly overshadows DNA barcoding, which is the approach currently conventionally used for species identification and widely used in ecological monitoring.
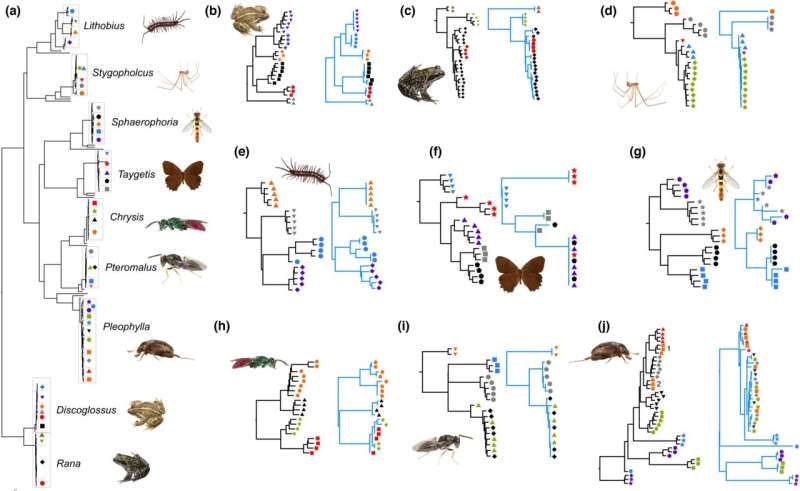
So far, USCOs have been used to check the completeness and quality of genome sequencing. The working group led by Dirk Ahrens, beetle researcher and collection curator at Museum Koenig, has now successfully used this method for the first time to separate cryptic species that were indistinguishable with conventional DNA barcodes.
The method was tested on spiders, centipedes, frogs, beetles, flies, small parasitic wasps, and butterflies. These new USCO markers can be obtained both by targeted sequencing and by extraction from genome data that has already been published and is available in databases. This makes the new approach sustainable and successful.
Although currently more expensive than conventional genetic barcoding, the authors propose metazoan USCOs primarily as the additional method of choice for the more complicated cases. There are quite a few of them, especially since science is still not even close to agreeing on how many species now inhabit Earth; estimates vary between 2 million and 2 billion species.
More information: Lars Dietz et al, Standardized nuclear markers improve and homogenize species delimitation in Metazoa, Methods in Ecology and Evolution (2022). DOI: 10.1111/2041-210X.14041
Journal information: Methods in Ecology and Evolution
Provided by Leibniz Institute for New Materials