Rationalized deep learning super-resolution microscopy for sustained live imaging of rapid subcellular processes

Li Dong's group from the Institute of Biophysics of the Chinese Academy of Science, in collaboration with DAI Qionghai's group from Department of Automation of Tsinghua University and Dr. Jennifer Lippincott-Schwartz from the Howard Hughes Medical Institute (HHMI) devised the rationalized deep learning super-resolution microscopy (rDL-SRM). The study was published in Nature Biotechnology.
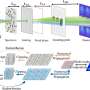
The researchers reported a method of rationalized deep learning super-resolution microscopy (rDL-SRM), which implants a deterministic physical model of specific microscopy into the network training and inference to eliminate/minimize the ill-posedness, model-uncertainty, spectral-bias, and demands for high-quality training dataset in current deep learning super-resolution (DLSR) methods.
They demonstrated that the rDL-SRM offers the first practical solutions for imaging a variety of dynamic intracellular processes at ultrahigh resolution in space and time for an unprecedented duration, including rapid kinetics inside motile cilium, the multi-phase transition of nucleolar proteins over light-sensitive mitosis, and long-time interactions between membranous and membrane-less organelles.
Instead of directly inferring the SR images as current super-resolution neural networks trained in an end-to-end manner, the rDL methods are applied to denoise the raw images, in which the high-frequency information beyond the diffraction limit is down-modulated as low-frequency Moiré fringes, and then reconstruct the SR image using the well-established SIM algorithm. This strategy eliminates the spectral-bias effect of recovering Moiré fringes, but also ensures the high-quality SR image reconstruction without spatial and temporal resolution degradation.
Systematic comparison demonstrated that the physical model rationalized three-branch architecture significantly improves the SR information by >10-fold and alleviates the model-uncertainty by 3~5-fold, providing physically feasible inferences that can be more readily generalized onto the regimes uncovered by training data.
The rDL concept is of wide compatibility and broad functionality. By utilizing the optical transfer function (OTF) of certain imaging modalities and the spatiotemporal continuity of noisy image series, the researchers constructed a learnable Fourier noise suppression module (FNSM) and the temporally/spatially interleaved self-supervised rDL (TiS/SiS-rDL) denoising networks, which can be implemented without the need to acquire ground-truth (GT) SR training data, but still yield results as good, sometimes even better, as the ones if supervised with GT, therefore, eliminating the need for extra training data.
rDL-SRM methods significantly push the boundaries of 2D/3D SR live-imaging significantly, achieving more than 30-fold longer imaging duration at a 10-fold higher frame rate than state-of-the-art SRM. Therefore, rDL-SRM methods enable investigating the fine spatial details, rapid kinetics, and long-time dynamics of a wide variety of bioprocesses, where SR live-imaging has not yet done so.
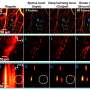
In a 2D SR imaging scenario, rDL TIRF-SIM achieved hour-long time-lapse super-resolution (97 nm) recording of cell spreading after the cells were placed on a coverslip, which clearly resolved the coordinated dynamics between F-actin and myosin-IIA, generating the protrusion and retraction oscillation of lamellipodia. The researchers utilized rDL GI-SIM to characterize the motile cilia beating pattern and frequency at SR imaging speed up to 684 Hz (limited by sCMOS camera read-out rate) for 60,000 frames without noticeable photobleaching for the first time, and the two-color live-cell visualization of cilium and IFT trains reveals multiple novel behavior of IFT trains colliding, remodeling, and turnaround in the midway of cilia, which was previously known of occurring only at the tip of cilia.
In a 3D imaging scenario, rDL LLS-SIM enables the researchers to analyze the multi-phase transition process of nucleoli over mitosis at an endogenous level. They consistently observe the large liquid droplets of RPA49 in the inner fibrillar center phase could partition into several independent foci, indicating that the active fission mechanism might play a role in the assembly of nucleoli. Moreover, TiS-rDL LLSM produced high-quality images of the collective behavior of transport SiT vesicles for 10,000 volumes at a high volumetric imaging speed of 10 whole-cell volumes per second. And SiS-rDL LLSM enables the hours-long three-color SR imaging at seconds interval, which first scrutinizes the diverse strategies cells employ to properly segregate different organelles during mitosis.
In conclusion, the rDL-SRM satisfies an unmet need for minimally invasive 2D/3D imaging of intracellular dynamics at ultra-high spatial and temporal resolution, high fidelity and quantifiability for long durations. The implementation and improvement of the rDL methods show great promise for shedding light on diverse biological phenomena.
More information: Chang Qiao et al, Rationalized deep learning super-resolution microscopy for sustained live imaging of rapid subcellular processes, Nature Biotechnology (2022). DOI: 10.1038/s41587-022-01471-3
Journal information: Nature Biotechnology
Provided by Chinese Academy of Sciences