Enzymatic reactions: Researchers reveal a regulatory mechanism by which life controls and organizes itself
Inside cells, molecular droplets form defined compartments for chemical reactions. Not only sticky interactions between molecules, but also dynamic reactions can form such droplets, as was found by researchers from the Max Planck Institute for Dynamics and Self-Organization (MPI-DS) and the University of Oxford. They revealed a new regulatory mechanism by which life controls and organizes itself.
Traditionally, cellular organelles defined by a membrane have been considered the functional units of a cell. In recent years, it has also been shown that molecular droplets formed inside the cell provide a micro-environment for important reactions. Such droplets are not enclosed by a membrane, and arise from phase separation. Hence, they form dynamically and can be regulated according to the needs of the cell.
Nonequilibrium drives can induce droplet formation
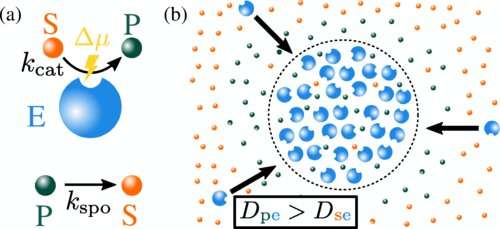
In the department of Living Matter Physics, managing director Ramin Golestanian and coworkers aim to reveal the organizational principles of living matter. "The formation of droplets in cells so far was ascribed to attractive, sticky interactions between molecules—similar to how droplets form in non-living, equilibrium systems, such as droplets of oil in a vinaigrette," explains Jaime Agudo-Canalejo, group leader at the MPI-DS.
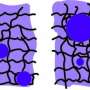
"We now found that the nonequilibrium drive provided by enzymatic reactions can cause the formation of enzyme-rich droplets, even without any stickiness. Instead, the enzymes are pushed against each other by the chemical fluxes they create," he continues.
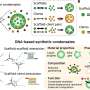
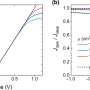
The researchers explored this novel mechanism by formulating a model in which the effect of a multicomponent enzymatic reaction on the micro-environment is described. They also considered the underlying feedback mechanism due to which the induced phase separation can in turn affect the initial enzymatic reaction.
"When the enzymatic activity gets too intense, phase separation occurs and acts to reduce it, providing a new form of autoregulation," says Matthew Cotton, first author of the study. This complex interplay of molecular interactions can provide a dynamic environment for cellular processes. Hence, the model adds another piece to the complex puzzle of how life is able to organize itself.
The research was published in Physical Review Letters.
More information: Matthew W. Cotton et al, Catalysis-Induced Phase Separation and Autoregulation of Enzymatic Activity, Physical Review Letters (2022). DOI: 10.1103/PhysRevLett.129.158101
Journal information: Physical Review Letters
Provided by Max Planck Society