Working to discover new treatments for tuberculosis
Mycobacterium tuberculosis, the causative agent of tuberculosis, remains the leading cause of infectious disease worldwide, affecting approximately a quarter of the globe's population. Treatment of infections is problematic due to the emergence of drug-resistant strains; however, University of Oklahoma professor Helen Zgurskaya, an expert in antibiotic resistance, is leading research on new potential therapeutic treatments for the disease.
Zgurskaya, a George Lynn Cross Research Professor in the Department of Chemistry and Biochemistry in the Dodge Family College of Arts and Sciences, is the corresponding author of the article "Proton transfer activity of the reconstituted MmpL3 is modulated by substrate mimics and inhibitors," published in Proceedings of the National Academy of Sciences.
"This is one of the scariest infectious diseases affecting billions of people worldwide," said Zgurskaya. "Like many other bacterial infections, it is becoming more resistant to antibiotics. Currently, the treatment requires a combination of antibiotics taken by patients for six months, but now imagine that the disease does not respond to the treatment. We are out of therapeutic options for this infection, and we need new drugs. The paper we published is focused on understanding how recently discovered new inhibitors kill the pathogen."
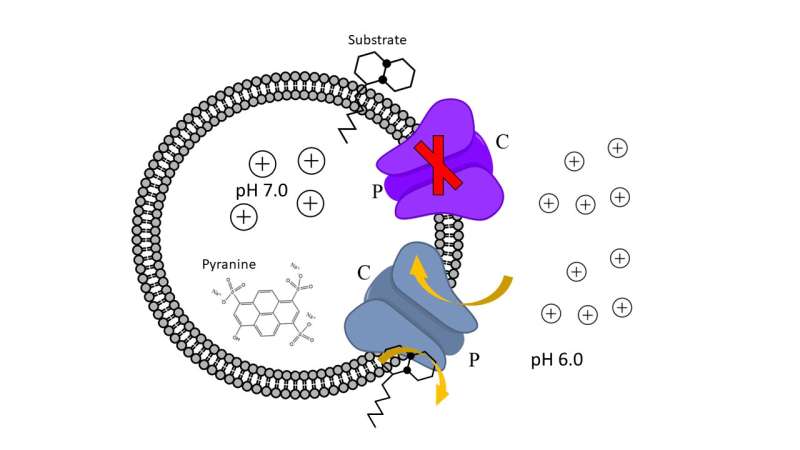
The team of researchers, which included Casey Stevens, Ph.D., postdoctoral research associate Svitlana Babii and research assistant professor Jitender Mehla, studied MmpL3 transporter and its analogs that are important for the physiology of Mycobacterium tuberculosis and antimycobacterial drug discovery. These transporters are critical for assembling the bacterial outer membrane that is needed for bacterial growth and resistance to antibiotics. In this study, researchers successfully purified and reconstituted MmpL3 and its analogs into artificial membranes. They also generated a series of substrate mimics and inhibitors specific to these transporters and analyzed their activities and properties.
Researchers found all reconstituted proteins facilitate proton translocation across membranes but the studied MmpL3 analogs differ dramatically in their responses to pH and interactions with substrate mimics and indole-2-carboxamide inhibitors. Their results further suggest that some inhibitors abolish the transport activity of MmpL3 and its analogs by inhibition of proton translocation.
The study provides a biochemical foundation for understanding the mechanism of these transporters and their inhibition by small molecule compounds that will facilitate the development of novel effective antibiotics.
Zgurskaya expects the next step would be to use the methods and techniques the team developed to analyze other inhibitors to identify those that are most effective, which will hopefully then go into clinical trials.
For this research, the OU team collaborated with scientists from Colorado State University, Creighton University and the Georgia Institute of Technology.
More information: Casey M. Stevens et al, Proton transfer activity of the reconstituted Mycobacterium tuberculosis MmpL3 is modulated by substrate mimics and inhibitors, Proceedings of the National Academy of Sciences (2022). DOI: 10.1073/pnas.2113963119
Journal information: Proceedings of the National Academy of Sciences
Provided by University of Oklahoma