Do proteins remember?
A study integrating biological ideas and new computer science tools has uncovered novel associations between genetic coding and protein structure, which could potentially change the way we think about protein production in the ribosome—the cell's "protein assembly line." The research, headed by Professor Alex Bronstein, Dr. Ailie Marx, and Ph.D. student Aviv Rosenberg, was published in Nature Communications.
Proteins, the complex molecules that play critical roles in virtually every biological mechanism, are produced by ribosomes in a process called translation. The ribosome decodes incoming "genetic instructions" to synthesize chains of amino acids—the building blocks of proteins. When amino acids are sequentially bound together into a long chain, they fold into a unique three-dimensional structure that grants the protein its biological properties and functionality. Errors in translation can lead to misfolding and subsequently physiological disorders, both mild and major.
Protein production instructions are delivered to the ribosome as codons, sequences of three "letters" from the genetic nucleotide code, which specify the identity and order of amino acids to be added by the ribosome to protein chain. For example, the codon UUU signals for addition of the amino acid phenylalanine, whereas codon UAC instructs for the addition of tyrosine. In this way, the codon sequence encodes for the unique sequence of amino acids characteristic to each protein. This mapping of genetic codons to amino acids used in translation is common to all living creatures on the planet, and is considered a primeval mechanism.
As if all of this were not complicated enough, it is important to point out that there are 61 codons that are decoded into just 20 amino acids. In other words, all but two amino acids are encoded by multiple codons.
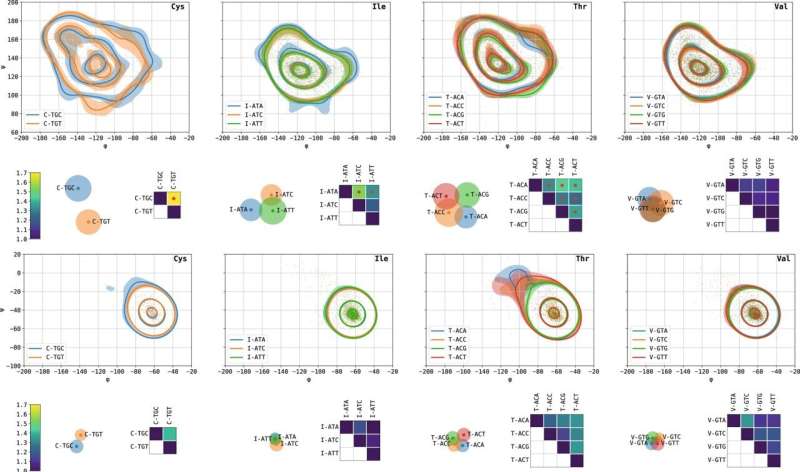
This is where the present research comes into the picture. Based on experiments carried out in the 1960s and 1970s, the accepted dogma states that proteins carry no "memory" of the specific codon from which each amino acid was translated as long as the amino acid identity remains unchanged. These early experiments into protein folding used chemical denaturants to unfold fully formed proteins and then demonstrated that upon removal of these chemicals the protein chain could refold spontaneously to regain its original structure and function. These experiments suggested that only the amino acid sequence, and not the specific codon sequence, determines a protein's structure. In view of this dogma, mutations that change the genetic coding without changing the amino acid are widely termed as "silent" and considered inconsequential for protein structure and function.
The Technion research team has uncovered an association between the identity of the codon and the local structure of the translated protein, which suggests that this may not be the general case and that proteins may indeed "remember" the specific instructions from which they were synthesized. The research team analyzed thousands of three-dimensional protein structures using dedicated tools they developed, which integrate advanced computer science methods, machine learning and statistics. In this way, they accurately compared the distributions of angles formed in these structures under different synonymous genetic codes. Their findings show that for certain codons, there is a significant statistical dependence between the identity of the codon and the local structure of the protein, at the position of the amino acid encoded by that codon.
The researchers emphasize that the findings are still unable to shed light on the direction of the causal relationship, meaning that it is not yet possible to say whether a change in genetic coding can cause a change in the local protein structure or whether structural changes may cause different coding, for example through evolutionary processes. This question is the foundation for a subsequent research study now being carried out by the group. According to Dr. Marx, a biologist by training and education, "If we find in subsequent research that the codon indeed has a causal effect on protein folding, this is likely to have a huge impact on our understanding of protein folding, as well as on future applications, such as engineering new proteins."
Dr. Marx emphasizes that the discovery presented in the article would not have been possible without Prof. Bronstein's computer and analysis skills. "This research is truly interdisciplinary, because biology alone cannot cope with such vast quantities of data without the help of data science, and computer scientists cannot themselves perform research of this kind, since they lack familiarity with the complex biological processes being probed. Therefore, our research highlights the huge advantage of interdisciplinary research that integrates skills from different fields to create a whole that is greater than the sum of its parts."
More information: Aviv A. Rosenberg et al, Codon-specific Ramachandran plots show amino acid backbone conformation depends on identity of the translated codon, Nature Communications (2022). DOI: 10.1038/s41467-022-30390-9
Journal information: Nature Communications
Provided by Technion - Israel Institute of Technology