Identification of enzyme involved in stress management in bacteria
Researchers from the University of East Anglia have identified a new class of enzyme involved in stress management in bacteria.
All organisms, from humans to bacteria, have to be able to respond to a wide range of stresses that result from changes in their environment.
Common amongst these are so called oxidative and nitrosative stresses, which occur when an organism is exposed to high concentrations of reaction oxygen or reactive nitrogen species, respectively. When this happens, fragile component of the cell are damaged, leading to loss of function and, in some cases, cell death.
Unsurprisingly, organisms have evolved a multitude of stress response systems that detect and alleviate particular stresses.
Iron-sulfur clusters, which consist of iron and inorganic sulfur, are found in all cell types where they play essential roles in a wide range of cellular processes. Because they are so reactive, they are often the first cellular components to become damaged under stress conditions.
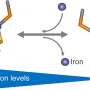
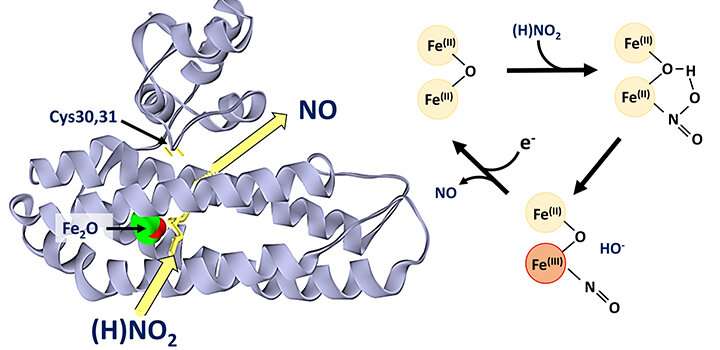
The di-iron protein YtfE, found widely in bacteria, is generally believed to function directly in the repair of iron-sulfur clusters that have been damaged under stress conditions. This activity has been variably proposed to involve donation of iron for re-building of iron-sulfur clusters, or the removal of nitric oxide (NO) from damaged clusters.
Recently, new evidence came to light from studies of YtfE function in cells that suggested its activity is associated with an increase, and not a decrease, in the concentration of NO. This prompted researchers in the School of Chemistry to re-examine the function of YtfE.
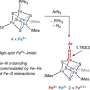
The team, led by Dr. Jason Crack and Prof Nick Le Brun, and involving Dr. Fraser MacMillan's lab, as well as researchers from the University of Birmingham and University of Sheffield, have discovered that YtfE is a new type of nitrite reductase enzyme that produces NO.
They showed that YtfE does not efficiently remove NO from damaged iron-sulfur clusters, nor is it an effective donor of iron for cluster assembly.
The YtfE-catalyzed production of toxic NO from nitrite (NO2-) may seem odd, but YtfE is co-regulated with another enzyme, called Hcp, which functions to detoxify NO (via its reduction to nitrous oxide, N2O).
The coupled YtfE/Hcp detoxification pathway represents an effective means by which the cell deals with toxic levels of nitrite that can occur under anaerobic conditions.
The team used a range of approaches, including in vivo genetic and in vitro protein spectroscopy and mass spectrometry, to gain detailed insight into the mechanism by which YtfE generates NO from nitrite.
The work clarifies the role of a protein that has puzzled researchers for a long time, demonstrating that YtfE is a key player for the management of stress under conditions of anaerobic respiration, such as those found in the human gut or when infecting a human host.
The work is published this week in the Journal of the American Chemical Society.
More information: Jason C. Crack et al, The Di-Iron Protein YtfE Is a Nitric Oxide-Generating Nitrite Reductase Involved in the Management of Nitrosative Stress, Journal of the American Chemical Society (2022). DOI: 10.1021/jacs.1c12407
Journal information: Journal of the American Chemical Society
Provided by University of East Anglia