Study finds eDNA 'game-changer' to help protect native animals
Curtin University researchers have identified a "game-changing" way of protecting native animals—including pygmy possums, western bush wallabies and Australian painted-snipe birds—using sophisticated DNA technology.
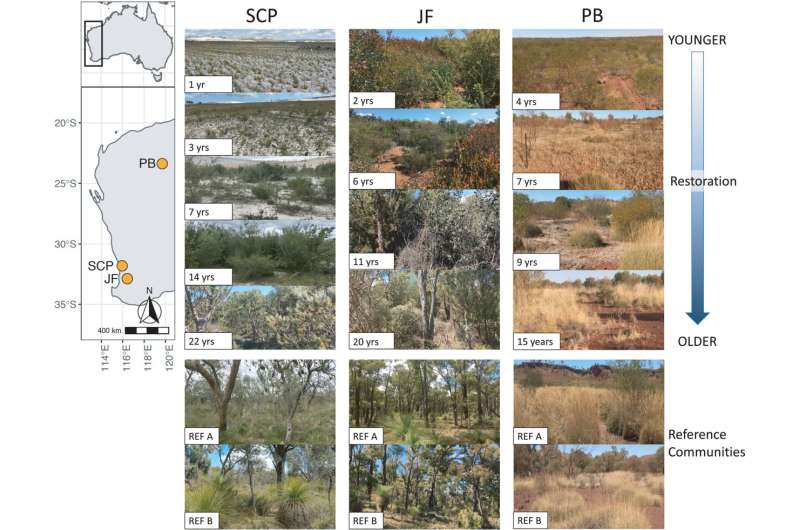
Two research papers, published in Molecular Ecology and Biodiversity and Conservation, examined animals and insects across multiple locations including Western Australia's Pilbara region, Perth region and the Jarrah Forest to find out where the use of DNA metabarcoding—a rapid DNA sequencing tool—would be most effective to monitor restoration.
Lead researcher Dr. Mieke van der Heyde, from Curtin's School of Molecular and Life Sciences, said DNA metabarcoding was a game-changer for monitoring the recovery of insects and animals because it could save time, money and resources.
"Fauna monitoring is often overlooked in restoration efforts because there is an assumption that as long as there are plants, everything else will come back on its own—and this isn't necessarily true," Dr. van der Heyde said.
"The problem is that monitoring fauna is hard, often requiring teams of experts in remote locations for weeks at a time, making it time-consuming and expensive. Tracking down the expertise to identify all the animals can also be difficult."
Dr. van der Hyde said DNA metabarcoding has a per sample cost rather than a per specimen cost, so it can be a cost-effective alternative to monitoring fauna recovery in diverse ecosystems but warned that it is not a 'one-size-fits-all' method.
"We found DNA metabarcoding can show the recovery of insects and plants in woodlands and forests, but not in the arid Pilbara region; and ground-dwelling insects give a better restoration signal than flying insects because they don't travel as far," Dr. van der Heyde said.
"In the arid Pilbara, we could tell the difference between restoration and reference sites using the animals detected from pooled poo samples. The lack of rain makes the droppings last longer and the lack of vegetation makes it much easier to see, especially bird droppings," Dr. van der Heyde said.
"Unfortunately, DNA metabarcoding can only identify animals and insects if we have reference DNA for them in our database. To improve this tool, we need to DNA barcode many more animals if we want accurate, species-level identifications from DNA.
"This technology can drastically improve conservation and restoration efforts for the many species under threat from loss of habitat and changing climates."
The papers are titled "Evaluating restoration trajectories using DNA metabarcoding of ground-dwelling and airborne invertebrates and associated plant communities" and "Scat DNA provides important data for effective monitoring of mammal and bird biodiversity."
More information: Mieke Heyde et al, Evaluating restoration trajectories using DNA metabarcoding of ground‐dwelling and airborne invertebrates and associated plant communities, Molecular Ecology (2022). DOI: 10.1111/mec.16375
M. van der Heyde et al, Scat DNA provides important data for effective monitoring of mammal and bird biodiversity, Biodiversity and Conservation (2021). DOI: 10.1007/s10531-021-02264-x
Journal information: Molecular Ecology
Provided by Curtin University




















