How cells defend against influenza A virus
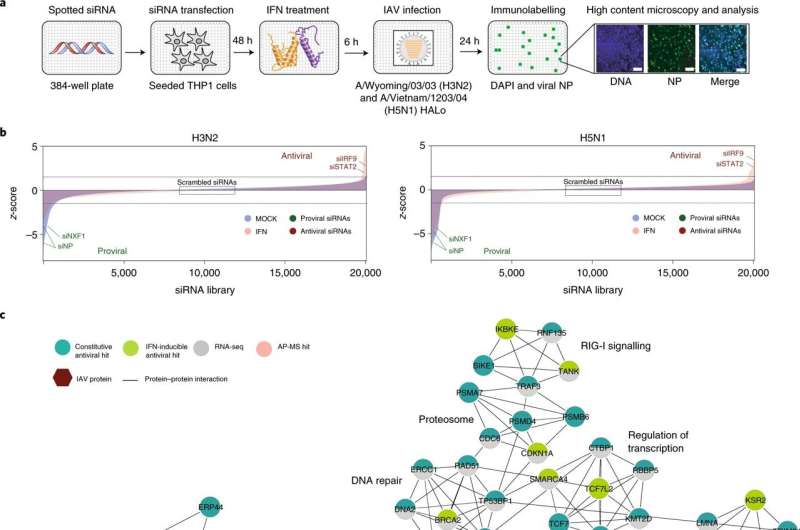
Human cells use a protein named TBC1D5 to route influenza A viruses inside host cells for destruction, preventing the virus from spreading replicated copies of itself to other cells, according to a study published in Nature Microbiology.
While influenza viruses have developed their own methods of overcoming this host defense, pharmacological methods to boost host defense systems could be one way to develop new anti-viral therapies, according to Judd Hultquist, Ph.D., assistant professor of Medicine in the Division of Infectious Diseases and a co-author of the study.
"This is one of the big dreams in virology, to identify host-targeted therapeutic strategies that limit the risk of antiviral resistance and have pan-viral potential," Hultquist said.
As far as viruses go, influenza A virus is extremely successful. Infecting more than 20 million people every year in the United States, the virus has developed sophisticated techniques to invade human cells and transform them into virus-producing factories. While human cells likewise develop mechanisms to prevent infection, viruses evolve and adapt much more quickly.
"We were very interested in learning about this arms race between virus and host," Hultquist said.
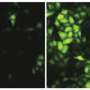
In the study, Hultquist's collaborators at the Sanford Burnham Prebys Medical Discovery Institute used a screening platform to knock-down almost every gene in the human genome and measure its effect on viral infection. When antiviral proteins are knocked down, they result in an increase in infection, which is how the investigators identified TBC1D5, Hultquist explained.
Investigators identified dozens of genes important to inhibiting viral replication, but TBC1D5 caught their attention given its known role in autophagy, a process of host cell protein recycling. When overexpressed, TBC1D5 interrupted the mechanism that transports new viruses from inside the cell to outside the cell, flagging the viruses as trash and instructing for them to be destroyed. However, in normal humans without overexpression of TBC1D5, this mechanism mostly fails.
This is because the virus actually has a counterattack: a protein that binds to the TBC1D5 "flag" and nullifies this host defense.
"The virus has devised an off switch for this recycling mechanism," Hultquist said. "Our cells have all of these amazing and clever defenses, but successful viruses have figured out how to 'one up' us and get around these challenges."
Identifying these critical interactions between host and pathogen are key to eventually developing anti-viral therapies that can help people fight off infections, such as a small-molecule inhibitor that nullifies the viral counterattack and boosts the efficacy of the TBC1D5 defense.
Furthermore, it is possible that many similar viruses use some of these same defensive strategies. For example, many respiratory viruses such as influenza viruses, SARS-CoV-2 and respiratory syncytial virus (RSV) all rely on the unique architecture of respiratory cells to replicate. It is possible that they have all developed unique ways to overcome this replication block, Hultquist said.
"If we can identify these common elements from virus to virus, we can begin developing these pan-viral therapies," Hultquist said.
More information: Laura Martin-Sancho et al, Restriction factor compendium for influenza A virus reveals a mechanism for evasion of autophagy, Nature Microbiology (2021). DOI: 10.1038/s41564-021-00964-2
Journal information: Nature Microbiology
Provided by Northwestern University