Genomic study shows abnormal DNA methylation in lead-exposed dogs
Epigenetics is the study of changes in gene activity which are not caused by changes in DNA sequences. DNA methylation—the addition of methyl groups to DNA—is the most well-known epigenetic modification. Lead exposure is known to cause epigenetic changes, and understanding these changes will help to more completely understand the impact and consequences of lead toxicity. This is especially important as epigenetic changes could affect multiple generations.
A team of researchers from Japan and Zambia, including Hokkaido University's Associate Professor Jumpei Yamazaki and Assistant Professor Shouta M. M. Nakayama, have identified abnormally high levels of DNA methylation in dogs exposed to high levels of lead near a mining area in Kabwe, Zambia. Their research was published in the journal Environmental Pollution.
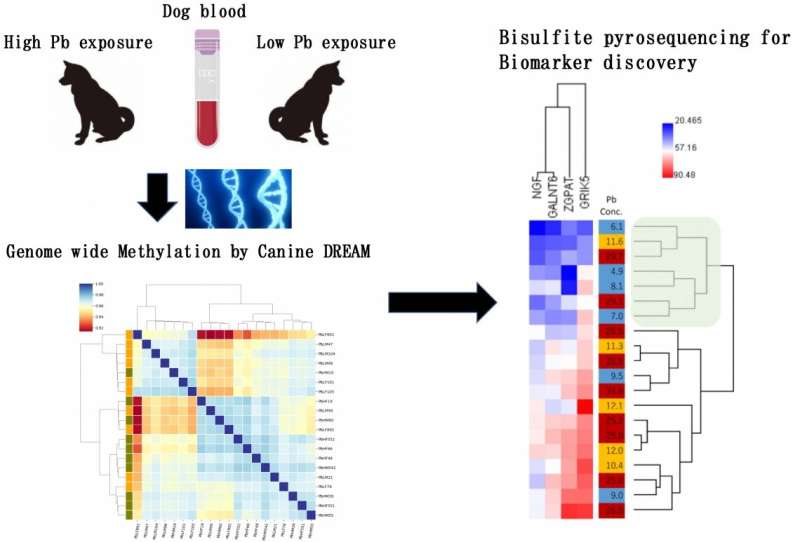
Between April and May 2017, the team took blood samples from 20 dogs whose lead concentration was examined using inductively coupled plasma mass spectrometry (ICP-MS), a highly sensitive analysis method. They decided to survey dogs in Kabwe, with the consent of their owners, because the dogs were more likely to be exposed to lead than humans. They discovered that the samples could be grouped into two clusters—one exposed to high levels of lead (an average lead concentration of 43.6 micrograms per deciliter of blood) and the other to low levels of lead (an average lead concentration of 7.2 micrograms per deciliter of blood).
From the blood samples, DNA was extracted and comprehensively analyzed by a next-generation sequencer using a method called Canine DREAM (digital restriction enzyme analysis of methylation) developed by the team, to discover changes in DNA methylation among the high- and low-exposure groups.
The analyses identified 827 sites in the dog genomes that exhibited differences in DNA methylation between and the low-exposure and high-exposure groups. Sites with increased methylation (hypermethylated sites) were mostly seen in the group of high-exposure dogs. Furthermore, 26 of these sites were associated with promoters, DNA sequences essential for the expression of genes, including a promoter for the gene involved in nerve growth. A validation study conducted on another group of 20 dogs in the same area showed a similar result.
The study suggests that hypermethylation, which is known to be associated with suppressed gene expressions, is highly affected by lead. It also validates previous studies that have shown that lead poisoning is linked to epigenetic alterations, especially DNA methylation.
The study suggests that similar epigenetic alterations could occur in humans and other animals. This could pave the way for the development of a new biomarker and treatment for lead-poisoning, a serious health problem seen around the world which causes symptoms including neurotoxicity, especially among babies and young children.
More information: Jumpei Yamazaki et al, Genome-wide DNA methylation analysis of dogs with high lead exposure living near a lead mining area in Kabwe, Zambia, Environmental Pollution (2021). DOI: 10.1016/j.envpol.2021.117229
Journal information: Environmental Pollution
Provided by Hokkaido University