Making sense of antisense gene silencing
Gene silencing therapies are used to interfere with, or 'silence', the expression of genes that are associated with disorders. Now, a team at TMDU has uncovered some of the cellular mechanisms by which the silencing therapies act in cells.
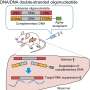
Antisense oligonucleotide (ASO) therapies use small strands of DNA or RNA that are antisense, or complementary, to the associated gene to interfere with its expression. ASO therapies are already available for some diseases, particularly neurological disorders, but their use is at a very early stage. It is known that modifying ASOs chemically can improve the efficacy of the therapy. The team at TMDU had previously achieved gene silencing by attaching alpha-tocopherol (Toc) to ASOs. They then created Toc-HDOs by attaching Toc to DNA/RNA heteroduplex oligonucleotides, which are double-stranded molecules consisting of one strand of DNA and one strand of RNA. Toc-HDOs are more potent, stable, and efficiently taken up by target tissues than ASOs, and so have great therapeutic potential.
However, very little is known about the mechanism by which Toc-HDOs function. ASOs are known to interact with proteins at every step of the gene silencing process, but none have been identified for Toc-HDOs. The team also wanted to investigate the possibility that the therapeutic effects of Toc-HDO might occur through a different mechanism to other ASOs, leading to the increased potency.
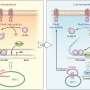
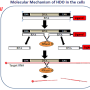
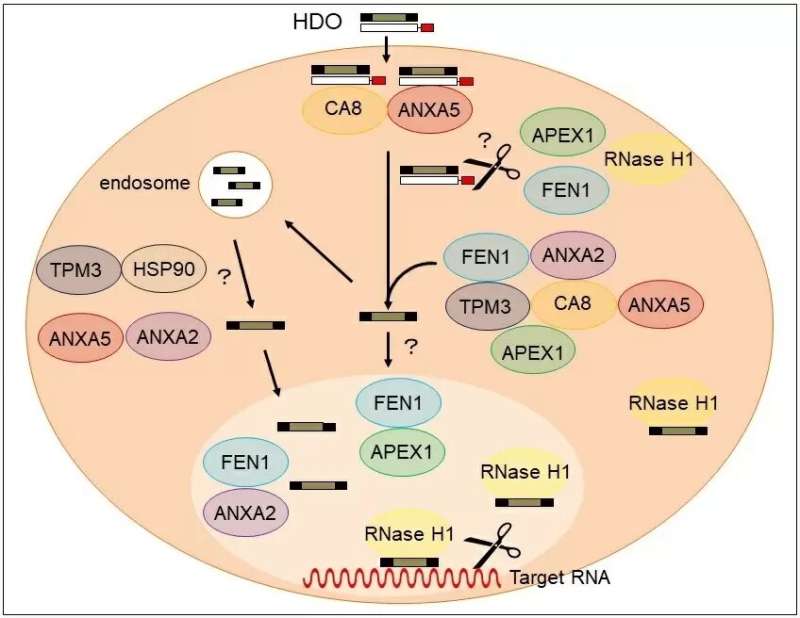
In this latest study, the researchers identified four proteins, annexin A5 (ANXA5), carbonic anhydrase 8 (CA8), apurinic/apyrimidinic endodeoxyribonuclease 1 (APEX1), and flap structure-specific endonuclease 1 (FEN1), all of which bind to Toc-HDO. "We injected mice with fluorescently-labeled Toc-HDOs to identify potential binding proteins," says lead author Ken Asada. "We then characterized the proteins further and showed that they are able to directly bind with Toc-HDOs in vitro."
The team demonstrated that these four proteins control the function of Toc-HDO and may bind together to form a complex to regulate gene silencing. "Tocopherol enhanced the binding activity of these proteins," says Takanori Yokota, senior author, "so this mechanism is a possible reason for the greater therapeutic potential shown by Toc-HDOs."
In this work, the researchers uncovered a novel biological mechanism, increasing understanding of how Toc-HDOs work to silence genes. This will allow the development of improved gene silencing therapies that are more potent and less toxic, advancing the field significantly.
More information: Ken Asada et al, Short DNA/RNA heteroduplex oligonucleotide interacting proteins are key regulators of target gene silencing, Nucleic Acids Research (2021). DOI: 10.1093/nar/gkab258
Journal information: Nucleic Acids Research
Provided by Tokyo Medical and Dental University