Microscopic imaging without a microscope? New technique visualizes all gene expression from a tissue sample
The 30,000 or so genes making up the human genome contain the instructions vital to life. Yet each of our cells expresses only a subset of these genes in their daily functioning. The difference between a heart cell and a liver cell, for example, is determined by which genes are expressed—and the correct expression of genes can mean the difference between health and disease.
Until recently, researchers investigating the genes underlying disease have been limited because traditional imaging techniques only allow for the study of a handful of genes at a time.
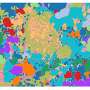
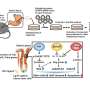
A new technique developed by Jun Hee Lee, Ph.D., and his team at the University of Michigan Medical School, part of Michigan Medicine, uses high-throughput sequencing, instead of a microscope, to obtain ultra-high-resolution images of gene expression from a tissue slide. The technology, which they call Seq-Scope, enables a researcher to see every gene expressed, as well single cells and structures within those cells, at incredibly high resolution: 0.6 micrometers or 66 times smaller than a human hair—beating current methods by multiple orders of magnitude.
"Whenever a pathologist gets a tissue sample, they stain it and look at it under the microscope—it's how they diagnose disease," explained Lee, an associate professor in the Department of Molecular & Integrative Physiology. "Instead of doing that, with our new method, we have made a microdevice that you can overlay with a tissue sample and sequence everything within it with a barcode with spatial coordinates."
Each so-called barcode is made up of a nucleotide sequence—the pattern of A, T, G, an C—found in DNA. Using these barcodes, a computer is able to locate every gene within a tissue sample, creating a Google-like database of all of the mRNAs transcribed from the genome.
"People have been trying to do this with other methods, such as microprinting, microbeads or microfluidic devices, but because of technological limitations, their resolution has been a distance of 20-100 micrometers. At that resolution you can't really see the level of detail needed to diagnose diseases," Lee said.
Lee adds that the technology has the potential to create an unbiased systematic way to analyze genes.
"Whenever we do science, we had to make a hypothesis about the role of two or three genes, but now we have genome-wide data at the microscopic scale and much more knowledge about what's going on inside that patient or model animal's tissue."
This knowledge could be used to provide insight into why certain patients respond to certain drugs while others do not, said Lee.
The team demonstrated the effectiveness of the technique using normal and diseased liver cells, successfully identifying dying liver cells, their surrounding inflamed immune cells and liver cells with altered gene expression.
"This technology actually showed many known pathological features that people have previously discovered but also many genes that are regulated in a novel way that was unrecognized previously," said Lee. "Seq-Scope technology, combined with other single cell RNA sequencing techniques, could accelerate scientific discoveries and might lead to a new paradigm in molecular diagnosis."
The work is described in the latest issue of the journal Cell.
More information: "Microscopic Examination of Spatial Transcriptome Using Seq-Scope," Cell, DOI: 10.1016/j.cell.2021.05.010
Journal information: Cell
Provided by University of Michigan