Researchers measure gene activity in single cells
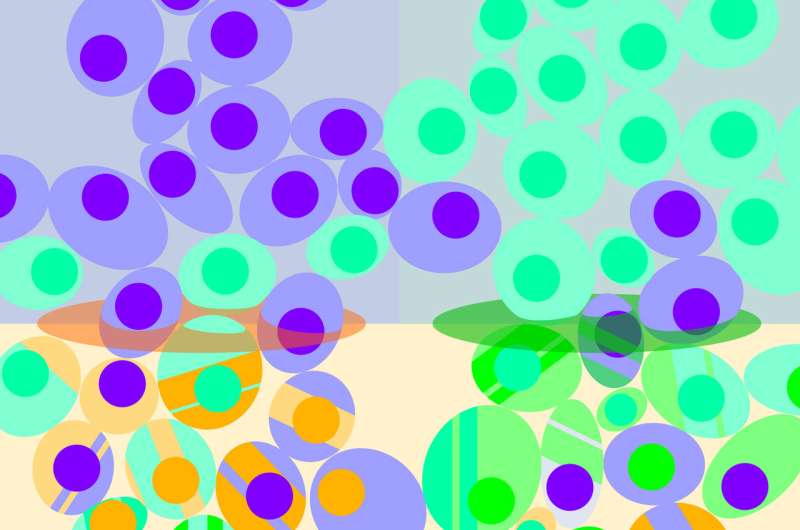
For biologists, a single cell is a world of its own: It can form a harmonious part of a tissue, or go rogue and take on a diseased state, like cancer. But biologists have long struggled to identify and track the many different types of cells hiding within tissues.
Researchers at the University of Washington and the Allen Institute for Brain Science have developed a new method to classify and track the multitude of cells in a tissue sample. In a paper published March 15 in the journal Science, the team reports that this new approach—known as SPLiT-seq—reliably tracks gene activity in a tissue down to the level of single cells.
"Cells differ from each other based on the activity of their genes—which genes are switched off or switched on," said senior author Georg Seelig, a UW associate professor in both the Department of Electrical Engineering and the Paul G. Allen School of Computer Science & Engineering. "Using SPLiT-seq, it becomes possible to measure gene activity in individual cells, even if there are hundreds of thousands of different cells in a tissue sample."
SPLiT-seq—which stands for Split Pool Ligation-based Transcriptome sequencing—combines a traditional approach to measuring gene expression with a new twist. For more than a decade, scientists have measured gene expression in tissues by sequencing the genetic "letters" of RNA, the DNA-like molecule that is the first step in gene expression. This standard approach—known as RNA-sequencing—profiles RNA across the whole tissue. But this approach does not tell researchers how cells within the tissue differ from one another. Single-cell RNA-sequencing addresses this by sequencing RNA from isolated cells, but existing methods are costly and do not scale well.
SPLiT-seq makes it possible to perform single-cell RNA-sequencing without ever isolating individual cells. The researchers put the cells through four rounds of "shuffling"—splitting them into separate pools and mixing them back together. At each shuffling step, they labeled the RNA in each pool with its own unique DNA "barcode." At the end of four rounds of shuffling and labeling, RNA from each cell essentially contained its own unique combination of barcodes—and that barcode combination is included in the bulk sequencing of all the RNA in the tissue.
"With these 'split-pool barcoding steps,' we solve a big problem in measuring gene expression: reliably identifying which RNA molecules came from which cell in the original tissue sample," said Seelig, who is also a researcher in the UW Molecular Engineering & Sciences Institute.
"With that problem addressed, we can begin to ask biological questions about the different types of cells we define in the tissue," said co-author Bosiljka Tasic, Associate Director of Molecular Genetics at the Allen Institute for Brain Science.
The team performed SPLiT-seq on brain and spinal cord tissue samples from laboratory mice. Using SPLiT-seq, they could measure the gene activity of over 156,000 cells. Based on patterns of gene activity, they estimated that more than 100 different types of cells were present in those tissue samples – including neurons and glial cells at various stages of development and differentiation.
SPLiT-seq can deliver this rich array of biological data at a cost of "just a penny per cell," said Seelig. This is a significantly lower cost than other single-cell RNA sequencing approaches, according to the researchers.
The researchers say that SPLiT-seq could answer important questions about how tissues develop, and identify minute changes in gene expression that precede the onset of complex diseases like Parkinson's disease or cancer.
More information: Alexander B. Rosenberg et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding, Science (2018). DOI: 10.1126/science.aam8999
Journal information: Science
Provided by University of Washington