Scientists discover how antibiotics target bacterial RNAP to inhibit its gene transcription
A group of researchers at the Hong Kong University of Science and Technology (HKUST) has uncovered the mechanism of how DNA is being melted to start bacterial gene transcription and how one class of antibiotics inhibits this process—an important way in killing bacteria. This discovery provides useful insight on the development of new antibiotics for bacteria that is antimicrobial resistance.
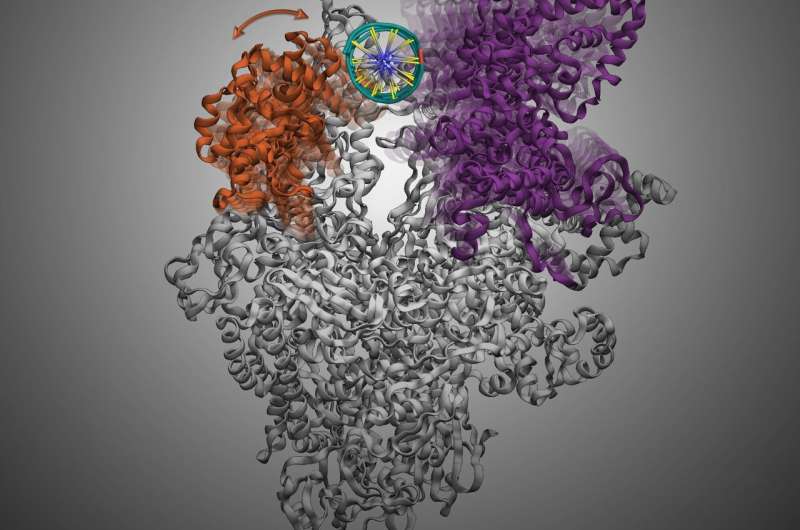
The emergence and spread of new forms of resistance remains a concern that urgently demand new antibiotics. Transcription is a vital process in bacterial cell, where genetic information in DNA is transcribed to RNA for the translation of proteins that perform cellular function. Hence, transcription serves as a promising target to develop new antibiotics because inhibition the transcription process should effectively kill the bacteria. Bacterial RNA polymerase, the core enzyme for transcription, must load the DNA and separate the double-stranded DNA to single stranded DNA to read the genetic information to initiate transcription. This process is also called DNA melting and is facilitated by the opening and closing of the loading gate of RNA Polymerase. The loading gate contains two flexible pincers (clamp and β-lobe) resembling the shape of a crab claw. DNA melting via this loading gate is a multi-step and highly dynamic process, and it provides a promising strategy for the design of novel antibiotics by inhibiting this process. Yet, the understanding of DNA melting requires a detailed understanding in the movements and dynamics of the loading gate, the lack thereof hampers future development of antibiotics.
To offer new direction for more effective therapeutics, a research team led by Prof. Xuhui Huang, Department of Chemistry and Department of Chemical and Biological Engineering at HKUST, recently discovered the working mechanism of an antibiotics, Myxopyronin, by targeting the movement of the loading gate to inhibit the DNA melting prior to bacterial gene transcription. The research team identified a partially closed form of the flexible clamp domain, into which an antibiotic called Myxopyronin can bind with. The binding of Myxopyronin to the RNA Polymerase diminishes the gate's ability to close, eventually inhibiting the DNA melting, which is vital for the survival of the bacteria.
More interestingly, the research team also found the unprecedented role of the β-lobe during the loading of DNA to the inner cleft of RNA Polymerase. They discovered that the opening of β-lobe is sufficient to accommodate the loading of double helix DNA without opening the Clamp. The role of β-lobe has not been previously reported, and this finding opens the opportunity to the development of new antibiotics targeting the β-lobe of RNA Polymerase to halt transcription.
"The shape of bacterial RNA polymerase resembles a crab claw that works like a pincer. The shape and flexibility of the two pincers are important for RNA Polymerase to hold and separate the double-helix form of DNA into single-stranded. We showed that an antibiotic that targets the movement of the pincers would be a promising as a drug candidate" said Prof. Huang. "What is more exciting, is that we also discovered a novel critical role of the β-lobe that can serve as a new target for future antibiotics development."
This work is made possible only with the quasi-Markov State Model (qMSM) recently developed in Prof. Huang's lab. qMSM is built from extensive all-atom molecular dynamics simulations, and successfully predicts dynamics of RNA Polymerase's loading gate at atomic resolution and millisecond (10-3 second) timescale. This new method adopts the generalized master equation formalism to encode non-Markovian dynamics, which has advantages over the popular Markov State Models based on Master Equation. Hence, it is especially promising to be applied to study complex conformational changes of proteins.
More information: Ilona Christy Unarta et al, Role of bacterial RNA polymerase gate opening dynamics in DNA loading and antibiotics inhibition elucidated by quasi-Markov State Model, Proceedings of the National Academy of Sciences (2021). DOI: 10.1073/pnas.2024324118
Journal information: Proceedings of the National Academy of Sciences
Provided by Hong Kong University of Science and Technology