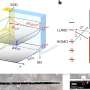
Algorithm maps gene expression in space
As we accumulate more and more gene-sequencing information, cell-type databases are growing in both size and complexity. There is a need to understand where different types of cells are located in the body, and to map their gene expression patterns into specific locations in tissues and organs. For example, a gene can be actively expressed in one cell while suppressed in another.
One way of mapping genes into tissues is a technique called in situ hybridization. Simply put, a target gene is tagged ("hybridized") with a fluorescent marker within the sections of the tissue it is located in (the "in situ" part). The sections are then visualized under a specialized microscope to see where the gene "lights up." Consecutive photographs of each section are then put together to generate a "spatial" map of the gene's location inside the tissue.
The problem with methods that use in situ hybridization is that, as the number of target genes grows, they start to become complicated, require specialized equipment, and force scientists to select their targets beforehand, a process that can be laborious if the goal is to reconstruct a full map of gene distribution across tissues.
"Spatializing" sequencing data
Now, scientists at EPFL's School of Life Sciences have created a computational algorithm called Tomographer, which can transform gene-sequencing data into spatially resolved data such as images, and does so without needing a microscope. The work was carried out by the research group of Gioele La Manno, and is now published in Nature Biotechnology.
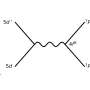
In the new Tomographer technique, the tissue is first cut along the axis of interest into consecutive sections, each of which is then sliced into tissue strips at different angles. Cells from the strips are then broken down to collect their total messenger RNA (mRNA). Each mRNA corresponds to a gene that was active in the cell. The measurements resulting from the strips can be then used as input to the Tomographer algorithm to reconstruct spatial gene-expression patterns across the tissue.
"The Tomographer algorithm opens a promising and robust path to "spatialize" different genomics measurement techniques," says Gioele La Manno. As an application, the scientists used Tomographer to spatially map the molecular anatomy of the brain of the Australian Bearded Dragon (Pogona vitticeps) – a non-model organism, demonstrating how versatile the algorithm can be.
"Ever since I started med school, I have been admiring the way computer tomography revolutionized the way we examine organs and body parts," says Christian Gabriel Schneider, one of the study's lead authors. "Today, I am very proud to be part of a team that has developed a molecular tomography technology. So far, we have focused on applications in neurodevelopmental biology, but in the future, we can certainly imagine molecular tomography becoming a constituent in personalized medicine."
"It was an exciting opportunity to develop an accessible and flexible computational method that has the potential to facilitate progress in the health sciences," adds Halima Hannah Schede, the study's other lead author. "I am very much looking forward to seeing what other spatially resolved biological data forms will be brought to light with Tomographer."
More information: Halima Hannah Schede et al. Spatial tissue profiling by imaging-free molecular tomography, Nature Biotechnology (2021). DOI: 10.1038/s41587-021-00879-7
Journal information: Nature Biotechnology
Provided by Ecole Polytechnique Federale de Lausanne