January 25, 2021 report
Using deep neural networks to identify features that may predict transcription factor binding
A team of researchers at the University of California, San Diego, has developed a deep neural network system to identify features that may predict transcription factor binding. In their paper published in the journal Nature Machine Intelligence, the group describes their system possible uses for better understanding transcription-factor-based diseases.
Transcription factors are proteins that play a role in controlling the rate of transcription of genetic information—the way they bind to DNA is the means by which genes are turned on or off. Prior research has shown that problems with transcription factors can lead to human diseases such as Rett syndrome, maturity-onset diabetes and Fuch's endothelial corneal dystrophy. Some research has suggested that they may also play a role in cancerous tumor development.
In order to prevent such diseases, scientists need to better understand the transcription process. In this new effort, the researchers built a neural-network-based system designed to assist with decoding the rules that govern transcription factors as they bind to target areas on strands of DNA. The team also hopes that it will prove useful in spotting specific noncoding nucleotides that have the biggest impact on binding.
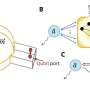
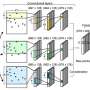
The team named their overall system framework AgentBind—it was built starting with a prior system developed at UCSD. The new system was made by putting together three convolutional neural networks, a connected layer and a combination recurrent and convolutional neural network. Because of the massive amounts of data involved in such research, the team used transfer learning (rather than bulk learning), making the learning process much more efficient. They also added a post-analytical process to generate importance scores to place bindings in context.
Testing involved running the system with transcription factors to see what it might yield—they found it was capable of providing new insights into transcription-factor-binding variants that might possibly be related to potential disease development. Such insights, they note, could lead to identifying what takes place when transcription factors go awry and cause diseases, which could potentially lead to the development of relevant therapies.
More information: An Zheng et al. Deep neural networks identify context-specific determinants of transcription factor binding affinity, bioRxiv (2020). DOI: 10.1101/2020.02.26.965343
An Zheng et al. Deep neural networks identify sequence context features predictive of transcription factor binding, Nature Machine Intelligence (2021). DOI: 10.1038/s42256-020-00282-y
Journal information: Nature Machine Intelligence
© 2021 Science X Network