Researchers reveal the first cryo-EM structures of NSD2 and NSD3 in complex with nucleosome
The nuclear receptor–binding SET Domain (NSD) family protein is closely connected with many cancers. However, their molecular mechanism remains unknown.
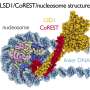
Recently, SUSTech research team made new achievements in the study of the cryo-EM structures of NSD2 and NSD3 in complex with nucleosome, which provided molecular insights into the nucleosome based recognition and histone-modification mechanisms of NSD2 and NSD3. The research results, titled "Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases' have been published in Nature.
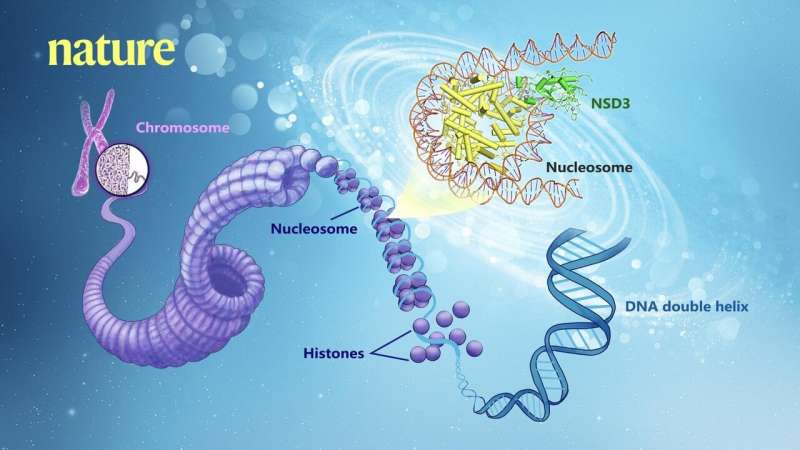
Genomic DNA wraps around the histone octamer (H2A, H2B, H3, and H4) to form the fundamental unit of chromatin–nucleosome. Histone lysine methyltransferases catalyze the transfer of up to three methyl groups to specific lysine (K) residues on the tails of histones H3 and H4, which is critical for the regulation of chromatin structure and gene expression. Dysregulation of histone lysine methyltransferases has been implicated in various cancers and numerous other diseases.
NSD family proteins are specific histone H3 lysine 36 methyltransferases. NSD2 plays a crucial role in the pathogenesis of multiple myeloma (MM) and pediatric acute lymphoblastic leukemia (ALL). Aberrant expression of NSD1 and NSD3 is also associated with a variety of human cancers, such as acute myeloid leukemia (AML), breast cancer, lung squamous cell carcinoma and squamous cell carcinoma of the head and neck. NSD methyltransferases exhibit an autoinhibitory state that is relieved by binding to nucleosomes, enabling di-methylation of histone H3 at Lys36 (H3K36). However, the molecular basis that underlies this mechanism remains unknown.
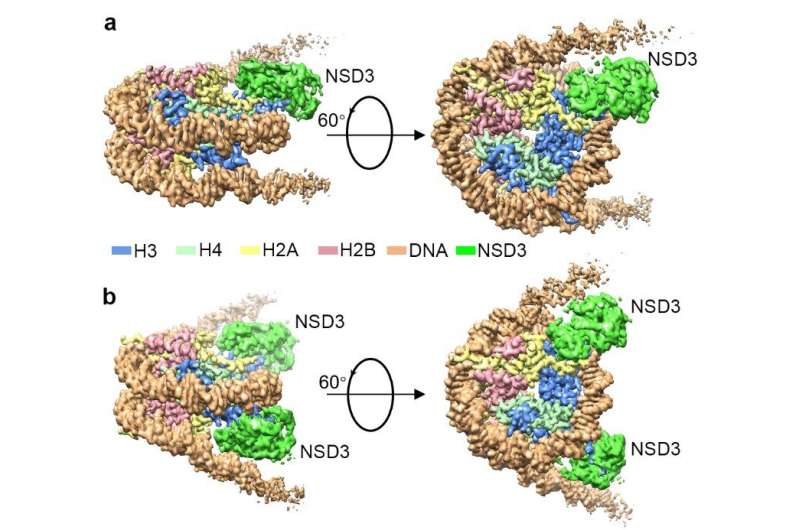
Researchers captured detailed three-dimensional molecular structures of NSDs protein in complex with nucleosome by state-of-the-art cryo-electron microscopy (cryo-EM). They have found that the binding of NSD2 and NSD3 to nucleosome causes DNA near the linker region to unwrap, which releases space for the insertion of NSD protein catalytic domain between the histone octamer and the unwrapped segment of DNA. The DNA- and histone-specific contacts between NSD2 or NSD3 and the nucleosome precisely position the enzyme on the nucleosome, explaining the H3K36 methylation specificity.
The researchers also found that the contacts between NSD proteins and nucleosomes are altered by the recurrent cancer-associated mutations in NSD2 and NSD3. These cancer-associated mutations promote cancer cell proliferation and xenograft tumor growth.
The molecular mechanism uncovered in this work will lead to the development of novel therapies and provide valuable information for the design and development of drugs for treating NSD-associated neoplastic diseases.
More information: Wanqiu Li et al. Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases, Nature (2020). DOI: 10.1038/s41586-020-03069-8
Journal information: Nature
Provided by Southern University of Science and Technology