Discovery opens new avenues for designing drugs to combat drug-resistant malaria

For the first time, UBC researchers have shown a key difference in the three-dimensional structures of a key metabolic enzyme in the parasite that causes malaria compared to its human counterpart.
The finding, recently published in the International Union of Crystallography Journal, brings researchers one step closer to developing new therapies to combat drug-resistant malaria.
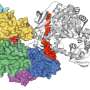
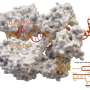
The structural differences of the metabolic enzyme, known as hexokinase, were captured using cryogenic electron microscopy or cryo-EM, whereby samples are cooled to cryogenic temperatures and examined at near-atomic resolution.
The researchers found that the enzyme in the malaria-causing parasite, known as Plasmodium, adopts a shape that has four individual subunits, while the enzyme in humans adopts a shape with just two subunits. This difference provides a unique opportunity to design drugs that specifically target the Plasmodium enzyme, without affecting the human version.
"Our new findings offer the prospect to design anti-malarial drugs that selectively target the unique structure of the parasite-infected cells, especially in the early stages of infection. Think of it as cutting a key based on the shape of the keyhole," says the study's senior author Dr. Sriram Subramaniam, the Gobind Khorana Canada Excellence Research Chair in UBC's faculty of medicine.
According to the World Health Organization, malaria, a mosquito-borne illness, affects more than 200 million individuals annually, resulting in the death of an estimated 400,000 each year. New treatments are in need, as resistance to the most common forms of anti-malarial drugs are on the rise, making the disease increasingly difficult to treat in some parts of the world.
"With this discovery, a new avenue for combatting drug-resistant malaria has opened," says the study's first author Dr. Shanti Swaroop Srivastava, a UBC postdoctoral fellow in Subramaniam's lab, which is recognized for world-leading contributions in cryo-EM. "Now that we know the atomic structural information of this key metabolic enzyme, new drugs can be developed to block the parasite's ability to metabolize glucose and survive."
The study, carried out in the laboratory of Dr. Subramaniam, was conducted in collaboration with research groups at Clemson University, led by Dr. James Morris, professor of genetics and biochemistry, and the Ohio State University Medical Center, led by Dr. Mark Drew, professor of microbial infection and immunity.
More information: Shanti Swaroop Srivastava et al, Plasmodium vivax and human hexokinases share similar active sites but display distinct quaternary architectures, IUCrJ (2020). DOI: 10.1107/S2052252520002456
Provided by University of British Columbia