April 9, 2020 feature
Sliding walls – a new paradigm for microfluidic devices
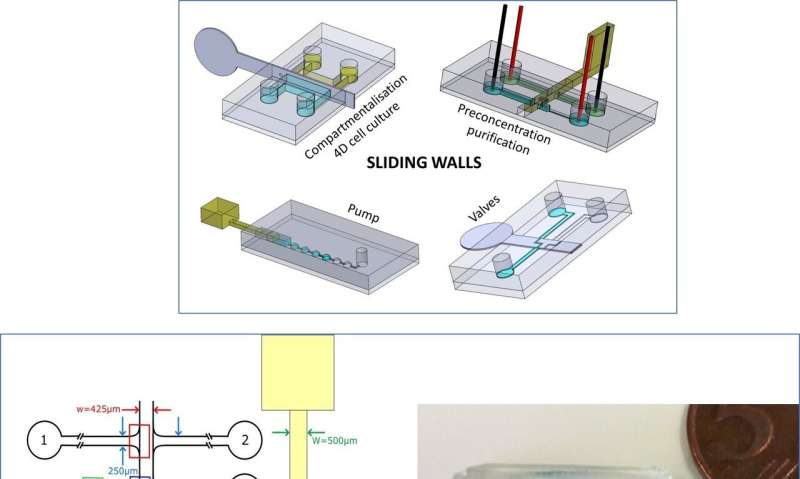
A research team recently developed "sliding walls" as a new technique for fluid control in microfluidic devices, allowing semi-rigid or rigid walls to slide inside a microfluidic chip. In a new report now on Nature: Microsystems & Nanoengineering, Bastien Venzac and a team of scientists at the Institute Curie and Sorbonne University in Paris, France, engineered several fluidic functions using sliding wall geometry. The device contained on/off switch valves to block or reconfigure channels depending on the wall geometry. The setup contained a hydrogel-based membrane to concentrate, purify and transport biomolecules from one channel to another. The technique is compatible with soft lithography methods for easy implementation based on typical fabrication workflows on polydimethylsiloxane (PDMS) chips. The new method opens a route to a variety of microfluidic applications, forming simple, hand-driven devices for point-of-care applications in biological labs.
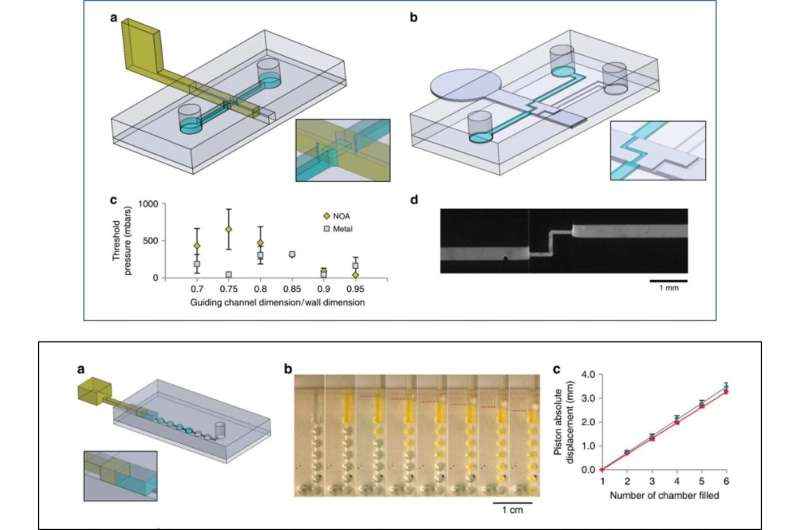
Truly reconfigurable systems are a microfluidics engineer's dream, where remodelling describes clever systems built in modular units and assembled for quick reorganization between experiments. For most microfluidic systems, however, the channel network remains fixed during microfabrication and cannot be custom restructured during the experiment. Engineers are also only able to conduct changes in pumping, valving or use external forces of electricity and magnetic fields. To meet the existing limits or challenges of microfluidic production, Venzac et al. proposed a new concept for microfluidic actuation known as "sliding walls". The method is compatible with soft-lithography fabrication but does not require external equipment. It can be manually operated and can be included into a single device component.
Venzac et al. developed sliding walls using several manufacturing methods to engineer them inside open-channels of polydimethylsiloxane (PDMS) chips. The actuation process allowed them to reversibly open or close a channel pumping fluids, then reorient flows to reconfigure a microfluidic network at will. The team described the principle of the method and demonstrated simple functions including formation of a hydrogel slab to accommodate four-dimensional (4-D), controlled cell culture, followed by membrane-based electrokinetic DNA preconcentration in microfluidic compartments. They implemented the technology at low cost for fast prototyping and manually controlled the sliding walls for simplicity, the team could also fully automate the walls using computer-controlled motors or actuators. The new toolbox is well adapted for applications with microfluidic channel dimensions above 100 µm and only require few actuation elements.
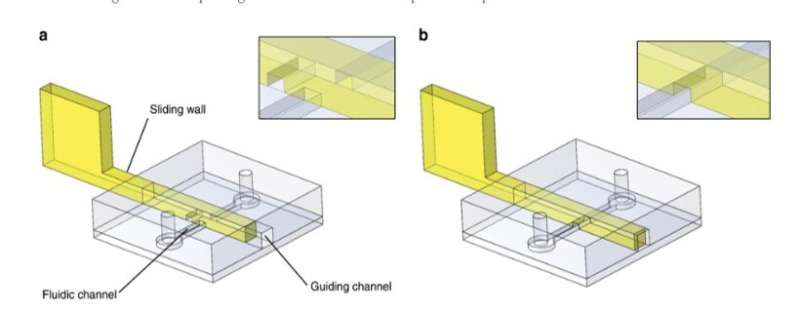
For the general design principle, the researchers inserted a rigid/semi-rigid structure into a guiding channel in the PDMS microfluidic chip and used a variety of materials to develop sliding walls including (1) stainless-steel films, (2) photocurable resist photopolymerized in PDMS moulds, and (3) photocurable resin moulded using stereolithographic 3-D printing. They selected the engineering techniques to fit the experiment according to their intrinsic properties and prevented wall buckling or breaking during actuation by controlling the material rigidity preferring stainless steel for most thin sliding walls. For larger sliding walls they used conventional stereolithography and used micro-milling on stainless steel to include small features on a sliding wall.
As an initial proof-of-concept, Venzac et al. prepared two types of valves: an on/off valve and a metallic switch valve with one inlet and two outlets. The sliding valves are mainly interesting due to their practicality in organ-on-chip devices and cell-culture constructs. The researchers also displayed the use of sliding walls as on-chip syringes to manually pump fluids and did not observe liquid leakage during pushing or aspiration of air in the experiments. The sliding walls were resourceful for large chamber construction—the team added two narrow grooves on the chamber roof and floor to guide a vertical stainless steel sliding wall and regulate communication between the compartments.
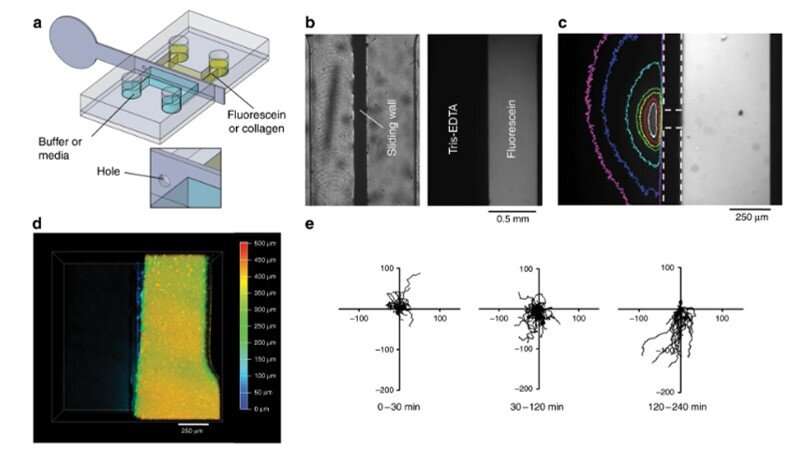
The team ultimately conducted biofunctionalization tests using the new device and observed 4-D cell culture and cell migration. In this experiment, they loaded a fluorescent collagen solution in the right half of the chamber, filled the second half with buffer and mixed the two to create a hydrogel slab. Such hydrogels are a major requirement to develop 3-D organ-on-chip compartments. To test their biological function, Venzac et al. studied cell migration with dendritic cells (immune cells) loaded in to the collagen solution inside a chamber. The team filled the second compartment with a chemokine solution and removed the stainless steel sliding wall to create a straight interface allowing the chemoattractant to diffuse onto the collagen slab for the dendritic cells to migrate onto the gel/solution interface, forming a 4-D cell culture.
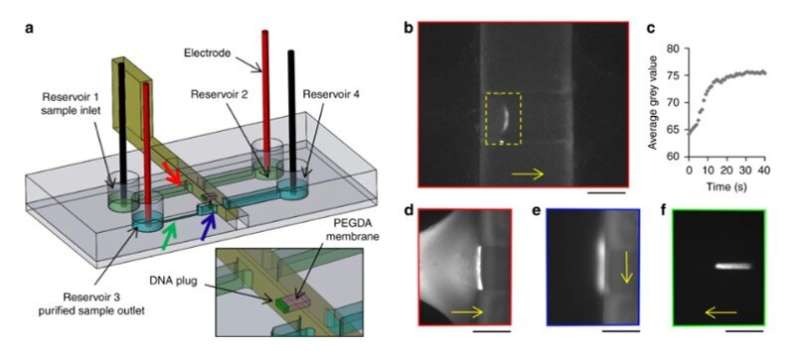
They also electrokinetically preconcentrated DNA macromolecules, controlled their transport and release in the new setup. To accomplish this, the team used a movable and reconfigurable hydrogel membrane in the microfluidic systems and engineered a sliding wall with an integrated window using high-resolution 3-D printing. They applied a constant electric field in the channels to allow electrophoretic migration of DNA-labelled with a fluorescent tag in buffer solution. The size of the hydrogel pores prevented DNA migration, causing them to preconcentrate at the membrane. The scientists induced free flow of preconcentrated DNA in the setup, to transport samples from one channel to another, as a new and simple route for sample preparation and analysis.
In this way, Bastien Venzac and colleagues developed a new toolbox to innovate the use of conventional microfluidics. The sliding walls had additional features such as microchannels or windows with loaded gels and solutions for potential applications beyond that of conventional in-chip valves. Notably, they achieved 4-D cell culture and DNA preconcentration using the single sliding wall setup. The scientists envision the technique in broad applications for low-cost and low-tech biomedical environments.
More information: Bastien Venzac et al. Sliding walls: a new paradigm for fluidic actuation and protocol implementation in microfluidics, Microsystems & Nanoengineering (2020). DOI: 10.1038/s41378-019-0125-7
Demetri Psaltis et al. Developing optofluidic technology through the fusion of microfluidics and optics, Nature (2006). DOI: 10.1038/nature05060
Yoojin Shin et al. Microfluidic assay for simultaneous culture of multiple cell types on surfaces or within hydrogels, Nature Protocols (2012). DOI: 10.1038/nprot.2012.051
Journal information: Nature , Nature Protocols
© 2020 Science X Network