How enzymes build sugar trees
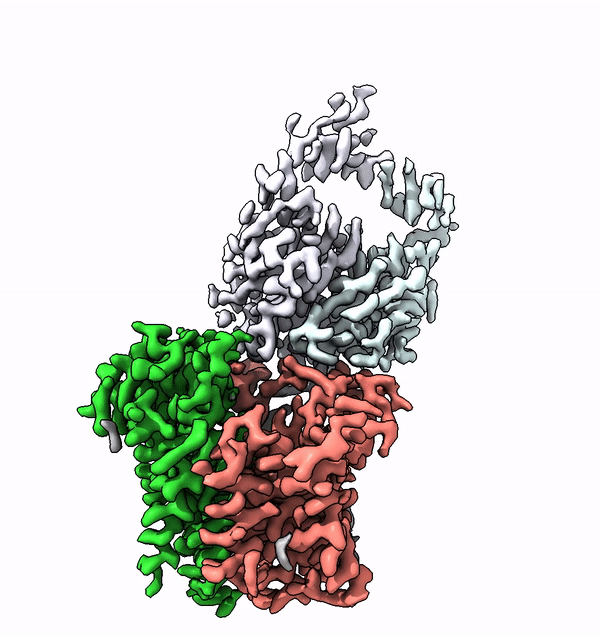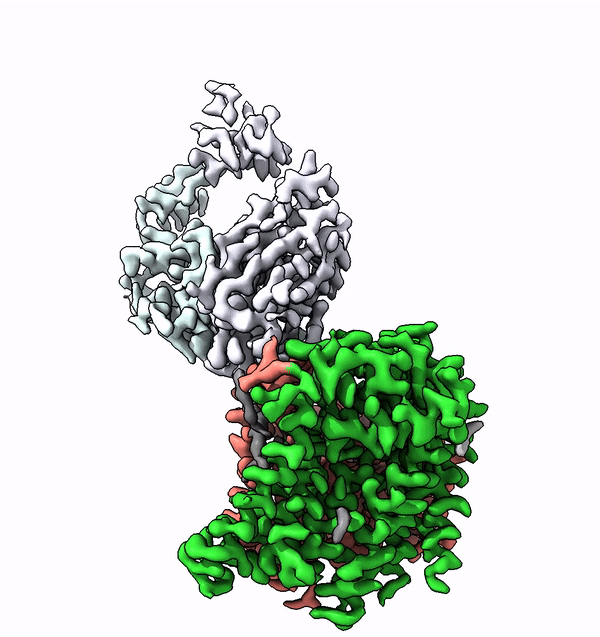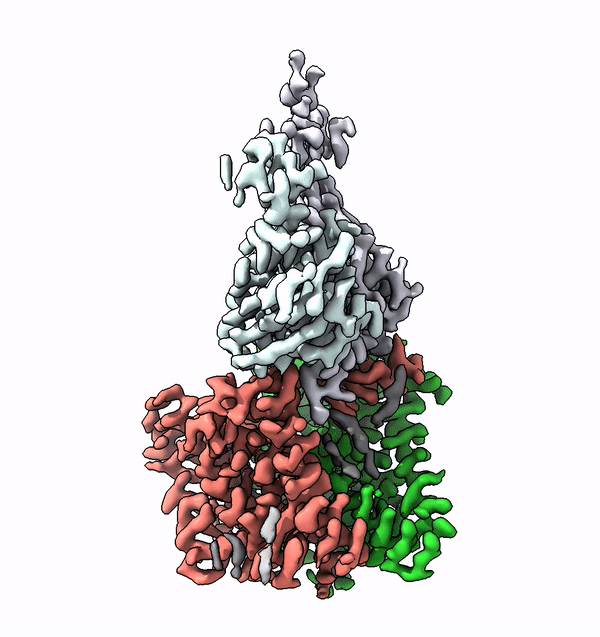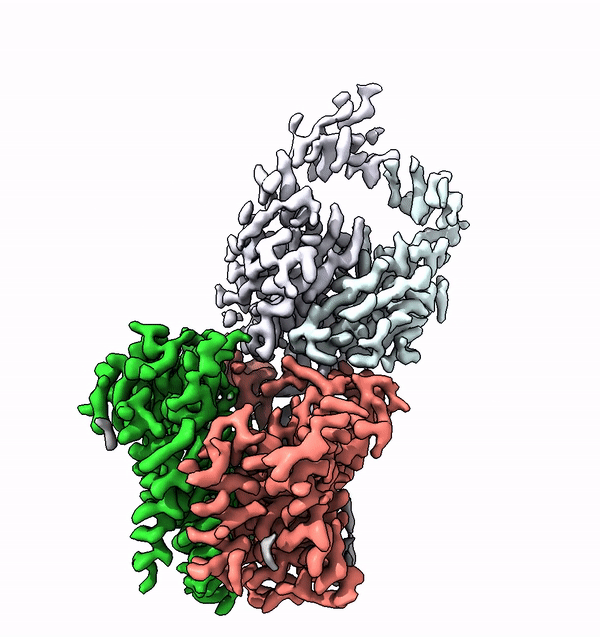
Researchers have used cryo-electron microscopy to elucidate for the first time the structure and function of a very small enzyme embedded in cell membranes. This enzyme builds complex sugar trees that are subsequently attached to other membrane proteins. The findings could accelerate the development of new, protein-based medications.
Many of the membrane proteins in eukaryotic cells are decorated with complex sugar trees called glycans. In addition to being extremely diverse, these sugar trees serve as a way to identify the respective organism, a cell type or its stage of maturity. For instance, the various blood groups in humans feature different glycans.
Complex sugars that are attached to lipids form a special class of glycans. In biology, these are known as lipid-linked oligosaccharides, or LLOs. LLOs are made up of a fat molecule embedded in the cell membrane and a sugar structure that extends either into the lumen of cell organelles or extracellularly.
Researchers from ETH Zurich, the University of Bern and the University of Chicago have now elucidated the structure of one of the enzymes responsible for the formation of LLOs. Their study has just been published in the latest issue of the journal Nature.
Modular protein architecture
The enzyme in question, known as ALG6, belongs to a superfamily of enzymes that the researchers call glycosyltransferases of the category C. Embedded in cell membranes, these link simple sugar molecules with other sugars in order to build sugar trees. They also link sugar molecules with proteins. In this capacity, glycosyltransferases play several key biological roles in all kinds of organisms, ranging from bacteria and fungi to highly developed mammals.
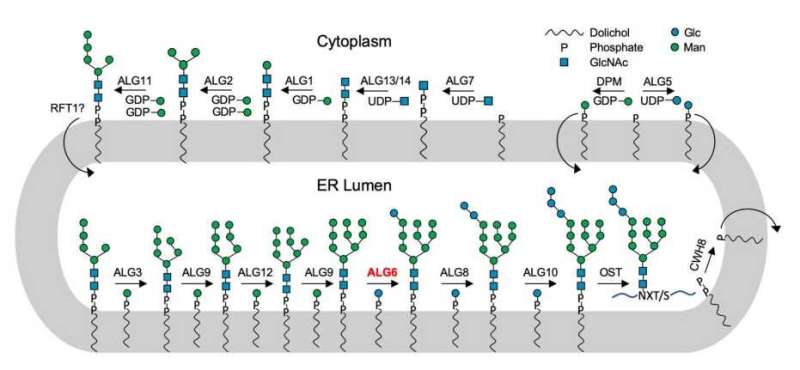
This enzyme superfamily had been a long-standing mystery to biologists because its individual members share hardly any structural motifs. The only things they do have in common is that they are membrane proteins that transfer sugars from one molecule to another, and that the sugars used for this transfer are always attached to lipids.
Based on the structure of ALG6, the group led by ETH Professor Kaspar Locher has now discovered that the members of this enzyme family have a modular design. Their research indicates that ALG6 and its relatives are made up of two modules: one whose structure is preserved during development, and a second, structurally variable module.
"We believe it's this modular design that helped these enzymes to evolve in different directions and, in turn, adapt to a large variety of different substrates," says Joël Bloch, an ETH doctoral student and the lead author of the study.
The findings finally explain the mechanism behind the enzyme family. "Our study has far-reaching implications for cell biology and for the production of therapeutic substances based on glycobiology," Bloch explains. These insights will be especially valuable in antibody engineering, which is currently of great interest to the pharmaceutical industry. They will also benefit the production of customised glycans in general, which are important for therapeutic proteins such as antibodies.
A record in cryo-electron microscopy
The researchers also see their results as a breakthrough in determining the molecular structures of proteins using cryo-electron microscopy (cryo-EM). In 2017, the Swiss researcher Jacques Dubochet received the Nobel Prize in Chemistry for his contribution to this groundbreaking technology, which has since become the method of choice for the structural elucidation of large molecular complexes.
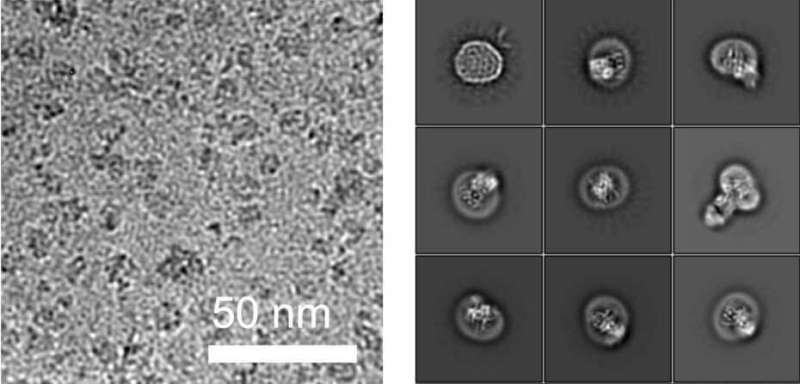
Determining the structures of small proteins at high resolution, especially those embedded in membranes, had not been possible using cryo-EM because measurements taken of particles below a certain mass do not permit precise structural calculations.
Working together with a research group from the University of Chicago, the ETH team have now found a solution to this problem. In collaboration with the Chicago-based researchers they produced a synthetic antibody that binds to ALG6. This antibody increased the mass of the ALG6 enzyme such that its structure could be determined in high resolution using cryo-EM.
"With our approach, we currently hold the world record for the highest structural resolution obtained for a membrane-bound complex of this size," Locher says with a hint of pride. He explains that these advances with cryo-EM will enable many other scientists to elucidate the structures of small membrane proteins: "Our approach paves the way for the scientific community to make rapid progress in the study of membrane proteins associated with a wide range of diseases."
A chemo-enzymatic toolbox
As if that were not enough, the ETH researchers, in collaboration with chemists from the University of Bern, have now developed methods for synthesising highly complex lipid-linked oligosaccharides in the lab—something that had not been possible with conventional synthesis methods in organic chemistry.
As a result, the researchers have now gained new insight into the essential cellular pathway of LLO biosynthesis, helping them explain how cells build complex glycans. "This marks a milestone in glycobiology that could form the basis for many glycobiologists' future research and for the production of glycoproteins," Locher says.
More information: Joël S. Bloch et al. Structure and mechanism of the ER-based glucosyltransferase ALG6, Nature (2020). DOI: 10.1038/s41586-020-2044-z
Journal information: Nature
Provided by ETH Zurich