Mining metagenomics: A faster and more efficient method to compare metagenomes
Microbiomes can contain thousands of bacteria species, hinting at the complex ecosystem that houses the microbiome as well as the one contained within the microbiome. Yet, until now, researchers have been limited in the tools they have to precisely compare those microbiomes.
Now, a team based in China has published a comprehensive comparison method for microbiomes in the December 3 issue of Bioinformatics, a journal of the Oxford University Press.
The researchers focused on improving the current method of examining the so-called "shotgun metagenomes." When an environmental sample is processed, all of the genetic material present is cut apart into identifiable DNA samples. The DNA samples are processed to produce metagenomes of the species in the environmental sample—a combination of metadata and genomes.
A metagenome can contain genetic data from several types of organisms, indicating the evolutionary combinations that lead to that specific species. High populations of specific organisms have a higher representation of the DNA sample, but even minor species can be accounted for through this method.
"An accurate and reliable distance (or dissimilarity) among shotgun metagenomes is fundamental to understanding microbiomes; however, current methods may produce erroneous results, due to their incomprehensive consideration of microbial compositions in comparing metagenomes," said paper author SU Xiaoquan from the Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT) of the Chinese Academy of Sciences (CAS).
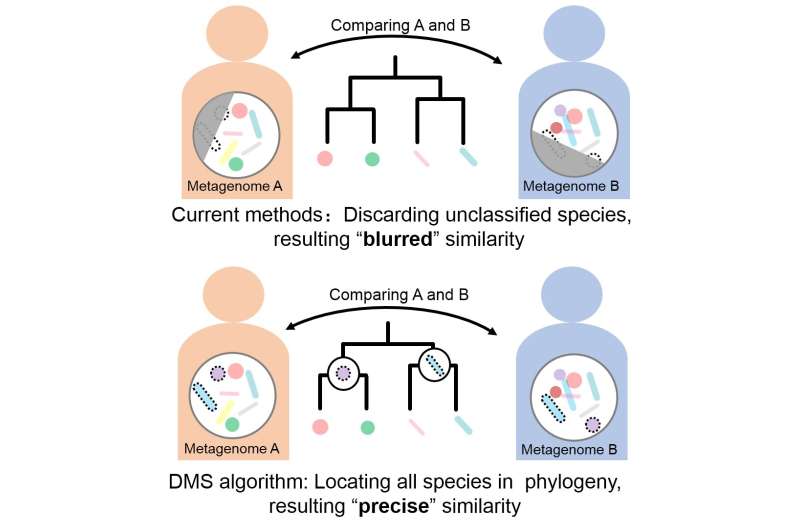
This incomprehensive consideration of microbial compositions includes discarding unclassified species or excluding evolutionary history or relationships among species.
To solve these problems, the researchers proposed the Dynamic Meta-Storms (DMS) algorithm. This method compares metagenomes—at the species level—for both their physical and genetic details, as well as their evolutionary relationships with other species.
"In addition, our new algorithm is also optimized for parallel computing: 20% faster than the currently most-used method and it also saves 40% memory," SU said. In 6.4 hours, the algorithm compared 100,000 metagenomes from 3,688 bacteria on a single computer node.
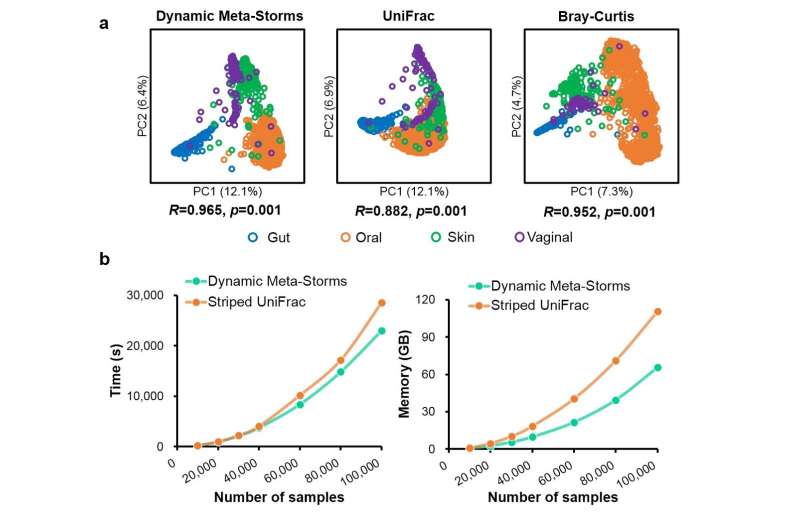
"The improved metagenomics comparison resolution and the high speed of the Dynamic Meta-Storms algorithm can be exploited to mine the massive quantity of metagenomes for classifying samples among various states, such as diagnosing the health status of a microbiome by comparing it to a reference database of diseased microbiomes," SU said.
The researchers made their tool available online for other scientists to use. They plan to continue improving the algorithm and working to identify metagenomes of microbiomes to better understand the evolutionary history of species.
More information: Gongchao Jing et al, Dynamic Meta-Storms enables comprehensive taxonomic and phylogenetic comparison of shotgun metagenomes at the species level, Bioinformatics (2019). DOI: 10.1093/bioinformatics/btz910
github.com/qibebt-bioinfo/dynamic-meta-storms
Journal information: Bioinformatics
Provided by Chinese Academy of Sciences