New strategies for optimizing the specificity of gene editing nucleases
Sangamo Therapeutics, Inc., a genomic medicine company, announced today the publication in Nature Biotechnology of a manuscript by Jeffrey Miller, Ph.D., and colleagues at Sangamo, describing two new strategies for optimizing the specificity of genome editing using zinc finger nucleases (ZFNs). The ability to engineer highly specific gene editing nucleases with little or no detectable activity at unintended genomic sequences is a key safety factor for therapeutic applications. The strategies entail engineering the two key functional regions within the ZFN structure, namely adjusting the binding affinity of the zinc finger array which recognizes DNA, and slowing the catalytic rate of the Fok1 cleavage domain. The two approaches, which are complementary, may be combined to enable near 100% on-target modification with no detectable off-targets. The manuscript, titled "Enhancing gene editing specificity by attenuating DNA cleavage kinetics," was published online on July 29 and will appear in the August issue of Nature Biotechnology.
"When attempting to improve the specificity of genome editing tools, on-target editing efficiency is often sacrificed," said Edward Rebar, Ph.D., Chief Technology Officer, Sangamo. "With these new strategies, the high efficiency we have observed previously with ZFN-mediated genome editing was completely preserved, while off-target activity was reduced by approximately 1000-fold, to below the level of detectability. These results are important as we believe the capabilities they demonstrate will help ensure the safety of our genome editing tools when used in the clinic."
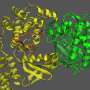
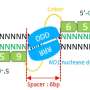
In order to adjust the binding affinity of the zinc finger recognition domain, the authors substituted a discrete, positively charged residue in the zinc finger framework to eliminate a nonspecific contact with the negatively charged phosphate backbone of the DNA. By varying the number of fingers bearing this substitution, the authors showed in cellular studies that they could effectively tune ZFN affinity into an optimally specific range, with no loss of on-target efficiency.
In a related series of cellular studies, the authors screened single-amino acid substitutions in the Fok1 nuclease domain in order to identify those able to improve specificity by slowing down catalysis. The goal of the study was to observe whether mutations would provide more time for the ZFNs to selectively dissociate from off-target sites prior to a cleavage event, which would improve global specificity. These studies yielded single-residue substitutions that could increase specificity by more than 1000-fold.
In a final study detailed in the manuscript, Sangamo scientists applied these two strategies in a therapeutically relevant setting by designing ZFNs that targeted the endogenous TCR-alpha gene in T-cells. Treatment of these T-cells with optimized ZFNs resulted in a greater than 98% on-target knockout efficiency of the TCR-alpha gene with undetectable off-target activity at a median assay background level of 0.01%. Sangamo believes these engineered improvements to the specificity of its ZFN genome editing platform have the potential to enable the routine generation of designed nucleases capable of high efficiency editing with minimal or no detectable off-target activity.
These results add to Sangamo's body of research demonstrating the high degree of precision, efficiency, and specificity of ZFNs for genome editing. In March 2019, Nature Communications published data demonstrating new ZFN architectures enabling high-precision genome editing. These new architectures yielded a 64-fold increase in the diversity of ZFNs available for targeting any DNA segment.
More information: Jeffrey C. Miller et al. Enhancing gene editing specificity by attenuating DNA cleavage kinetics, Nature Biotechnology (2019). DOI: 10.1038/s41587-019-0186-z
Journal information: Nature Biotechnology , Nature Communications
Provided by Sangamo Therapeutics, Inc.