An effective sweeper closes DNA replication cycling
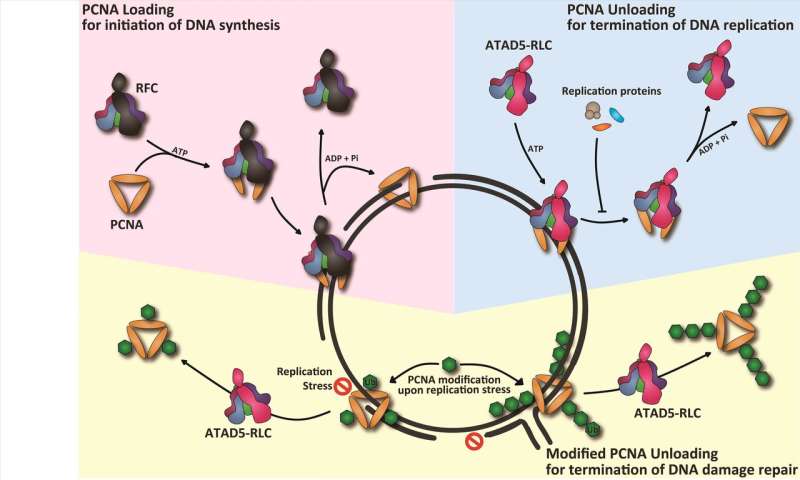
DNA replication is essential for living organisms to faithfully deliver genetic information from parental cells to daughter cells. Many proteins are assembled on the parental DNA to work as replication machineries. Among them, the proliferating cell nuclear antigen (PCNA) is a key replication protein. This ring-structured molecule encircles chromosomes, thread-like structures where DNA molecules is packed into, during DNA replication. Just like a ring on a string, PCNA-ring is tightly linked to DNA. After its landing on DNA, PCNA recruits other proteins to efficiently copy the parental DNA. PCNA's stable connection to DNA makes it an essential platform for many replication machineries.
This is, however, not the end of the story. When the DNA synthesis is complete, PCNA should be removed from the DNA to ensure a complete end of its cycling. Otherwise, the replication proteins make genomic DNA unstable that may lead to genetic mutations. How then are replication machineries cleared from DNA after DNA replication? PCNA unloading from DNA clears replication machineries from DNA after DNA replication. Although PCNA unloading is crucial for maintaining genomic stability, it has been remained unclear how PCNA is unloaded during replication termination.
Director Myung Kyungjae, professor Kim Hajin and Dr. Kang Sukhyun at the Center for Genomic Integrity within the Institute for Basic Science (IBS) at the Ulsan National Institute of Science and Technology (UNIST), South Korea, reported a novel molecular mechanism for the regulation of PCNA cycling during DNA replication. They found that a specific protein complex called ATAD5-RFC-Like-Complex (ATAD5-RLC) is a PCNA unloader. Dr. Myung's group has established an in vitro reconstitution system by which PCNA loading and unloading could be monitored. The ATAD5-RLC allows the PCNA ring to be removed from DNA as a bona-fide PCNA unloader.
These findings come from their previous studies. Another protein complex, Replication-Factor-C (RFC) is known to open the PCNA-ring and load it onto a linear DNA molecule before DNA synthesis. Until now, scientists in the DNA replication field believed RFC also function to unload PCNA, since a bacterial homolog of RFC does both loading and unloading reactions. Dr. Myung's group suggested that ATAD5-RLC, having a similar structure to RFC, could be a candidate for a PCNA unloader. This study proved biochemically that ATAD5-RLC is a bona-fide PCNA unloader. The scientists also revealed that ATAD5-RLC could unload modified PCNA, too. PCNA is modified upon replication stress to deal with DNA damage. These findings highlight the role of ATAD5-RLC as a universal sweeper of PCNA for the termination during DNA replication and repair.
The scientists also identified key motifs in ATAD5 that make the protein complex a PCNA unloader. They revealed the mechanistic differences between PCNA loading and unloading processes. Additionally, the scientists found a point mutation in a key motif in ATAD5 from melanoma patients. Dr. Sukhyun Kang, one of the corresponding authors of the study says, "It was crucial to functionally dissect ATAD5 and purify active ATAD5-RLC. Establishment of in vitro reconstitution system allowed us mechanistic studies for PCNA unloading."
This study provides a complete understanding of PCNA cycling on replicating DNA. It has not been well understood which complex is responsible for the unloading of PCNA after DNA replication and repair. The scientists show that two structurally related complexes play distinct roles in PCNA-DNA association. RFC loads PCNA to initiate DNA synthesis and ATAD5-RLC unloads PCNA to terminate DNA replication/repair. Mechanistic differences between loading and unloading of PCNA revealed by this study offer a conceptual advance to understanding DNA replication. Since PCNA unloading is closely linked to chromatin assembly, this study will be the basis of future studies for the relationship between replication termination and maintenance of epigenetic information.
"This is a major advance to understanding regulatory mechanisms for the replication termination process. Since uncontrolled disassembly of replication machineries causes genomic instability that may result in cellular transformation, our study will be beneficial to develop strategies for cancer treatment," explains director Myung.
More information: Mi-Sun Kang et al. Regulation of PCNA cycling on replicating DNA by RFC and RFC-like complexes, Nature Communications (2019). DOI: 10.1038/s41467-019-10376-w
Journal information: Nature Communications
Provided by Institute for Basic Science



















