A new sequencing method to detect DNA modifications of relevance to cancer

Ludwig Cancer Research scientists report in the current issue of Nature Biotechnology a new and improved method to detect chemical modifications to DNA. These modifications—or "epigenetic" marks—help control gene expression and their aberrant distribution across the genome.
Led by Chunxiao Song and Benjamin Schuster-Boeckler, both assistant members of the Ludwig Institute for Cancer Research, Oxford, the study demonstrates that their method, dubbed TET-assisted pyridine borane sequencing, or TAPS, is a less damaging and more efficient replacement for bisulfite sequencing, the current gold standard for mapping epigenetic modifications to DNA.
"We think TAPS could directly replace bisulfite sequencing as a new standard in DNA epigenetic sequencing," said Song. "It makes DNA epigenetic sequencing more affordable and accessible to a wider range of academic research and clinical applications."
One class of epigenetic modifications involves the attachment of chemical groups to one of the four "letters," or bases, of DNA. These marks do not alter the DNA sequence itself, but rather influence the switching on and off of genes. Two of the most common modifications of this type involve the addition of chemical methyl and hydroxymethyl groups to cytosine to create 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC). "Abnormal patterns of 5mC and 5hmC abundance are classic hallmarks of cancer," Song said.
For decades, biologists have relied on bisulfite to detect 5mC and 5hmC modifications, but the chemical is extremely destructive, degrading as much as 99 percent of the DNA it contacts. "You can imagine that if you're dealing with very limited genetic samples, such as cell-free DNA circulating in blood, bisulfite sequencing becomes very challenging," Song said.
Another limitation of bisulfite sequencing is that it detects 5mC and 5hmC only indirectly. The technique converts unmodified cytosine to a related base called uracil, while leaving methylated cytosine intact. "This is very inefficient and computation intensive," Song said. "It's like looking for Waldo by eliminating everyone who is not Waldo. TAPS allows us to find Waldo directly."
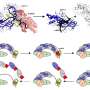
The new method involves two steps. The first uses an enzyme known as TET to convert 5mC and 5hmC to a third modification, 5-carboxylcytosine (5caC). The second step involves a new chemical reaction developed by Yibin Liu, a postdoctoral researcher in Song's lab, to convert 5caC into thymine—a DNA base that can be read by ordinary sequencing machines.
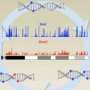
The process generated a new type of data, and the laboratory of Schuster-Boeckler developed the computational methodology required for its analysis. "Compared to bisulfite sequencing, processing TAPS data is not only twice as fast, it also retains a lot more information from the original sample. This makes it much easier to detect mutations and structural variations even while identifying DNA modifications," Schuster-Boeckler said.
After demonstrating that the core chemistry of TAPS worked, the team spent another year perfecting it, pushing the efficiency of the reaction from about 70 percent to over 95 percent. The researchers also developed two variations of the TAPS technique—TAPS-Beta and CAPS—which can be used to detect only 5mC or 5hmC, respectively.
In tests with mouse DNA, the researchers showed that TAPS can generate more accurate sequencing data at half the cost of bisulfite sequencing. "We can get twice as much useful data for the same amount of money," Song said.
Song, Schuster-Boeckler and colleagues are now refining TAPS to analyze cell-free DNA shed by tumors into the bloodstream with the goal of applying the technique to develop minimally invasive cancer diagnostics.
More information: Yibin Liu et al, Bisulfite-free direct detection of 5-methylcytosine and 5-hydroxymethylcytosine at base resolution, Nature Biotechnology (2019). DOI: 10.1038/s41587-019-0041-2
Journal information: Nature Biotechnology
Provided by Ludwig Institute for Cancer Research