Physicists find the limits of multitasking in biological networks

Many complex systems in biology can be conceptualized as networks. This perspective helps researchers understand how biological systems work on a fundamental level, and can be used to answer key questions in biology, medicine, and engineering.
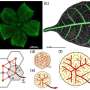
Blood flow in the brain is a prime example. Blood travels through a network of vessels and can be re-routed to specific parts of the brain as needed. Walking, for example, would require blood flow in different regions than chewing gum.
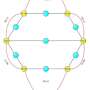
It's thought that networks perform such tasks by controlling connections within the network, called "edges." What physicists hadn't explored is how many tasks a single network can accomplish simultaneously.
A team of researchers from the Department of Physics & Astronomy published a study in PNAS that addresses this question. Graduate student Jason W. Rocks and former postdoc Henrik Ronellenfitsch, who is now at MIT, were the lead authors of this paper, and worked alongside physicists Andrea Liu and Eleni Katifori, as well as Sidney R. Nagel from the University of Chicago.
The Penn team had previously studied two types of networks. Katifori has examined how nature builds and maintains "flow networks," such as blood flow, using approaches that are inspired by and related to biology. Liu studies "mechanical networks," such as the arrangement of amino acids that form a protein, and how these networks can be changed in order to perform a specific biological function.
While these two systems differ from one another, discussions between the Liu and Katifori groups about how much multitasking each network could accomplish helped Liu and Katifori realize that they could study these two seemingly unrelated networks together.
"We were both independently studying the complexity of a particular function that a flow network could do and what a mechanical network could do," says Katifori. "It was two entirely different physical networks, but in a way the same question."
The authors developed a set of equations that described each system. They then used simulations to control or "tune" the network so they would perform increasingly complex functions. Rocks, Ronellenfitsch, and their colleagues found that both types of networks succeeded at multitasking.
They were surprised by the similarities in performance between these two seemingly distinct networks. While the physics underlying the two systems is entirely different, they performed similarly in terms of multitasking abilities and controllability. "Quantitatively, they were almost identical," says Liu.
These results will serve as the foundation for a number of future studies that will delve deeper into how the ability to perform tasks is encoded into networks. For mechanical networks like enzymes, this knowledge could improve biomedical researchers' ability to design targeted drugs and treatments.
As a first step, Rocks is working on better understanding how the networks actually work. "Up to this point we've treated it like a black box," he says. "But we don't want to do that. We want to understand how a network performs a specific function. We want to understand what aspects of the network's structure are important."
Liu and Katifori are enthusiastic about their collaboration and the results they hope to find in the near future. "If you had asked me before we did this project whether we were going to have the same answer for the two networks, I would say 'why?'" Katifori says. "But then when you think about it, and when you understand it, you realize the elegance of this study and why these two networks should be the same."
More information: Jason W. Rocks et al, Limits of multifunctionality in tunable networks, Proceedings of the National Academy of Sciences (2019). DOI: 10.1073/pnas.1806790116
Journal information: Proceedings of the National Academy of Sciences
Provided by University of Pennsylvania