Evolution sans mutation discovered in single-celled archaea

University of Nebraska-Lincoln researchers have found revolutionary evidence that an evolutionary phenomenon at work in complex organisms is at play in their single-celled counterparts, too.
Species most often evolve through DNA mutations inherited by successive generations. A few decades ago, researchers began discovering that multicellular species can also evolve through epigenetics: traits originating from the inheritance of cellular proteins that control access to an organism's DNA, rather than genetic changes.
Because those proteins can respond to shifts in an organism's environment, epigenetics resides on the ever-thin line between nature and nurture. Evidence for it had emerged only in eukaryotes, the multicellular domain of life that comprises animals, plants and several other kingdoms.
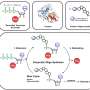
But a series of experiments from Nebraska's Sophie Payne, Paul Blum and colleagues has shown that epigenetics can pass along extreme acid resistance in a species of archaea: microscopic, single-celled organisms that share features with both eukaryotes and bacteria.
"The surprise is that it's in these relatively primitive organisms, which we know to be ancient," said Blum, Charles Bessey Professor of Biological Sciences at Nebraska. "We've been thinking about this as something (evolutionarily) new. But epigenetics is not a newcomer to the planet."
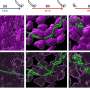
The team discovered the phenomenon in Sulfolobus solfataricus, a sulfur-eating species that thrives in the boiling, vinegar-acidic springs of Yellowstone National Park. By exposing the species to increasing levels of acidity over several years, the researchers evolved three strains that exhibited a resistance 178 times greater than that of their Yellowstone ancestors.
One of those strains evolved the resistance despite no mutations in its DNA, while the other two underwent mutations in mutually exclusive genes that do not contribute to acid resistance. And when the team disrupted the proteins thought to control the expression of resistance-relevant genes—leaving the DNA itself untouched—that resistance abruptly disappeared in subsequent generations.
"We predicted that they'd be mutated, and we'd follow the mutations, and that would teach us what caused the extreme acid resistance," Blum said. "But that's not what we found."
Though epigenetics is essential to some of the most productive and destructive physiological processes in humans—the differentiation of cells into roughly 200 types, the occurrence of cancers—it remains difficult to study in eukaryotes.
The simplicity of archaea, combined with the fact that their cells resemble eukaryotes' in some important ways, should allow researchers to investigate epigenetic questions much faster and more cheaply than was possible before, Blum said.
"We don't know what flips the switch in humans that changes epigenetic traits," Blum said. "And we sure don't know how to reverse it very often. That's the first thing we'll go after: how to turn it on, how to turn it off, how to get it to switch. And that has benefits when you think about (managing) traits in us or traits in plants."
Yet the discovery also raises questions, Payne said, especially about how both eukaryotes and archaea came to adopt epigenetics as a method of inheritance.
"Maybe both of them had it because they diverged from a common ancestor that had it," said Payne, a doctoral student in biological sciences. "Or maybe it evolved twice. It's a really interesting concept from an evolutionary perspective."
Blum said the team is likewise curious about whether and how epigenetics might explain why no known archaea cause disease or wage antibiotic-armed warfare against their brethren, as bacteria do.
"There are no antibiotics going on in that world," he said. "Why is that? We're thinking (that) it's got something to do with epigenetics, and so their interactions among each other are fundamentally different than bacteria."
The research introduces an even broader question, Blum said.
"What was the benefit for them to have this? We don't know."
The team reported its findings in the journal Proceedings of the National Academy of Sciences.
More information: Sophie Payne et al. Nonmutational mechanism of inheritance in the Archaeon Sulfolobus solfataricus, Proceedings of the National Academy of Sciences (2018). DOI: 10.1073/pnas.1808221115
Journal information: Proceedings of the National Academy of Sciences
Provided by University of Nebraska-Lincoln