AI heralds new frontiers for predicting enzyme activity
Researchers from the Departments of Chemistry and Engineering Science at the University of Oxford have found a general way of predicting enzyme activity. Enzymes are the protein catalysts that perform most of the key functions in Biology. Published in Nature Chemical Biology, the researchers' novel AI approach is based on the enzyme's sequence, together with the screening of a defined 'training set' of substrates and the right chemical parameters to define them.
Enzymes are the target of many drugs. If scientists can predict their functions, they can then inhibit those functions with small molecules—in some cases to treat disease. This research will be critical to creating an holistic picture that is provides a fuller and more complete understanding of biology and health.
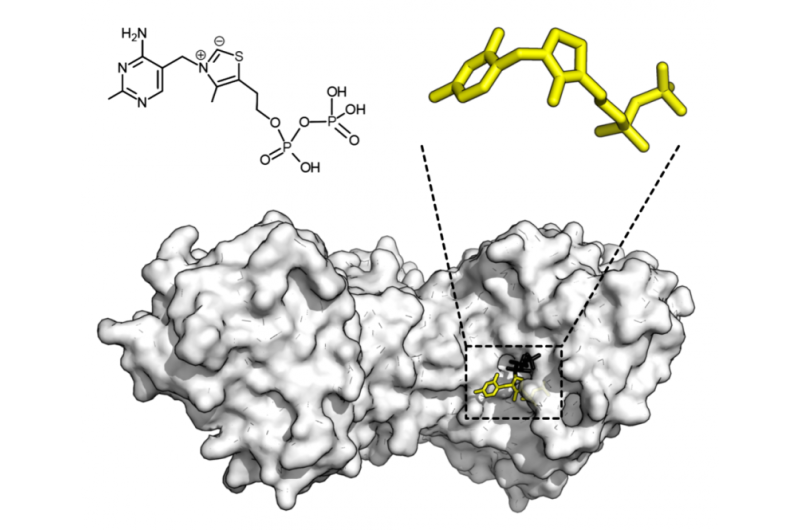
The researchers tackled an entire family of enzymes from one plant species. They combined high-throughput expression of the enzymes from the corresponding genes, then screened their enzymatic activity by quantitative, label-free mass spectroscopy. Simple analysis of the enzyme's primary sequence gives no real pattern of activity prediction, but when combined with AI techniques from Oxford University's Machine Learning Group, standard chemical descriptors can derive a powerfully predictive system.
Ben Davis, Professor of chemistry at the University of Oxford says, "The key thing is that rather than being 'black box' this method gives back to the chemist/biologist successful predictions and reasons for those predictions that have chemical and biological meaning. This in turn has allowed us to work out which enzymes can be used in synthesis, predict the activity of enzymes from very different species (even bacteria) and to work out how to engineer enzymes in a new way based on suggestions that we wouldn't have predicted."
He adds: "We see this as being a very powerful discovery engine. It will throw intriguing possibilities into the mix for hypothesis testing. Given the recent chemistry Nobel Prize in the test tube evolution of enzymes, AI applied to enzymes for increased understanding could prove to be a very powerful next frontier."
Stephen Roberts, professor of machine learning in information engineering at the University of Oxford says: "We live in an era of big data and big models, but not necessarily of big knowledge or insight. Indeed, the nature of many complex, well performing models obscures the details of success, leading to 'black-box' solutions which lack ready interpretability. In sharp contrast, the scientific method builds insight extraction into its core. In this research we have shown that models that provide transparency and insight are still capable of driving scientific advances."
This major advance enables successful protein catalyst activity predictions, which has implications a huge range of areas including medical research. It is a significantly more challenging field than modelling small molecule catalysts which has been the zenith in machine learning/chemistry until now.
More information: Min Yang et al. Functional and informatics analysis enables glycosyltransferase activity prediction, Nature Chemical Biology (2018). DOI: 10.1038/s41589-018-0154-9
Journal information: Nature Chemical Biology
Provided by University of Oxford