Genes – way weirder than you thought
Pretty much everyone, at least in societies with access to public education or exposure to media in its various forms, has been introduced to the idea of the gene, but "exposure does not equate to understanding" (see Lanie et al., 2004). Here I will argue that part of the problem is that instruction in genetics (or in more modern terms, the molecular biology of the gene and its role in biological processes) has not kept up with the advances in our understanding of the molecular mechanisms underlying biological processes (Gayon, 2016).
Let us reflect (for a moment) on the development of the concept of a gene: Over the course of human history, those who have been paying attention to such things have noticed that organisms appear to come in "types", what biologists refer to as species. At the same time, individual organisms of the same type are not identical to one another, they vary in various ways. Moreover, these differences can be passed from generation to generation, and by controlling which organisms were bred together; some of the resulting offspring often displayed more extreme versions of the "selected" traits. By strictly controlling which individuals were bred together, over a number of generations, people were able to select for the specific traits they desired (→). As an interesting aside, as people domesticated animals, such as cows and goats, the availability of associated resources (e.g. milk) led to reciprocal effects – resulting in traits such as adult lactose tolerance (see Evolution of (adult) lactose tolerance & Gerbault et al., 2011). Overall, the process of plant and animal breeding is generally rather harsh (something that the fanciers of strange breeds who object to GMOs might reflect upon), in that individuals that did not display the desired trait(s) were generally destroyed (or at best, not allowed to breed).
Charles Darwin took inspiration from this process, substituting "natural" for artificial (human-determined) selection to shape populations, eventually generating new species (Darwin, 1859). Underlying such evolutionary processes was the presumption that traits, and their variation, was "encoded" in some type of "factors", eventually known as genes and their variants, alleles. Genes influenced the organism's molecular, cellular, and developmental systems, but the nature of these inheritable factors and the molecular trait building machines active in living systems was more or less completely obscure.
Through his studies on peas, Gregor Mendel was the first to clearly identify some of the rules for the behavior of these inheritable factors using highly stereotyped, and essentially discontinuous traits – a pea was either yellow or green, wrinkled or smooth. Such traits, while they exist in other organisms, are in fact rare – an example of how the scientific exploration of exceptional situations can help understand general processes, but the downside is the promulgation of the idea that genes and traits are somehow discontinuous – that a trait is yes/no, displayed by an organism or not – in contrast to the realities that the link between the two is complex, a reality rarely directly addressed (apparently) in most introductory genetics courses. Understanding such processes is critical to appreciating the fact that genetics is often not destiny, but rather alterations in probabilities (see Cooper et al., 2013). Without such an more nuanced and realistic understanding, it can be difficult to make sense of genetic information.
A gene is part of a molecular machine: A number of observations transformed the abstraction of Darwin's and Mendel's hereditary factors into physical entities and molecular mechanisms. In 1928 Fred Griffith demonstrated that a genetic trait could be transferred from dead to living organisms – implying a degree of physical / chemical stability; subsequent observations implied that the genetic information transferred involved DNA molecules. The determination of the structure of double-stranded DNA immediately suggested how information could be stored in DNA (in variations of bases along the length of the molecule) and how this information could be duplicated (based on the specificity of base pairing). Mutations could be understood as changes in the sequence of bases along a DNA molecule (introduced by chemicals, radiation, mistakes during replication, or molecular reorganizations associated with DNA repair mechanisms and selfish genetic elements.
But on their own, DNA molecules are inert – they have functions only within the context of a living organism (or highly artificial, that is man made, experimental systems). The next critical step was to understand how a gene works within a biological system, that is, within an organism. This involve appreciating the molecular mechanisms (primarily proteins) involved in identifying which stretches of a particular DNA molecule were used as templates for the synthesis of RNA molecules, which in turn could be used to direct the synthesis of polypeptides (see previous post on polypeptides and proteins). In the context of the introductory biology courses I am familiar with (please let me know if I am wrong), these processes are based on a rather deterministic context; a gene is either on or off in a particular cell type, leading to the presence or absence of a trait. Such a deterministic presentation ignores the stochastic nature of molecular level processes (see past post: Biology education in the light of single cell/molecule studies) and the dynamic interaction networks that underlie cellular behaviors.
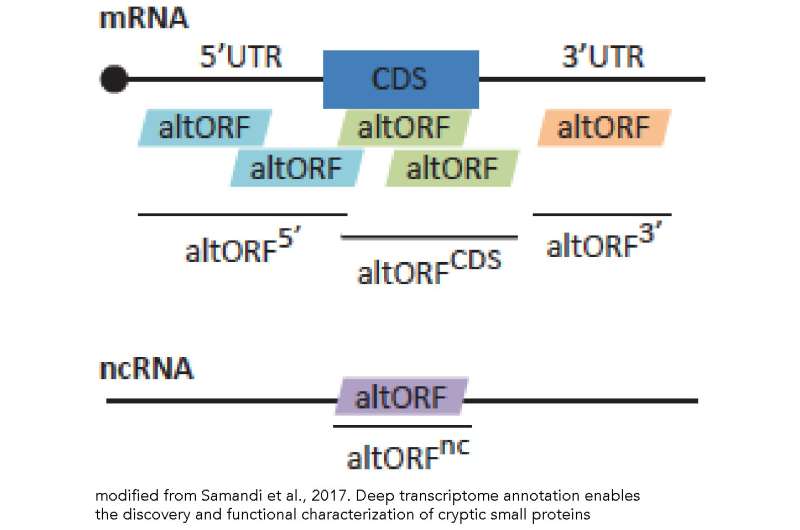
But our level of resolution is changing rapidly. For a number of practical reasons, when the human genome was first sequence, the identification of polypeptide-encoding genes was based on recognizing "open-reading frames" (ORFs) encoding polypeptides of > 100 amino acids in length (> 300 base long coding sequence). The increasing sensitivity of mass spectrometry-based proteomic studies reveals that smaller ORFs (smORFs) are present and can lead to the synthesis of short (
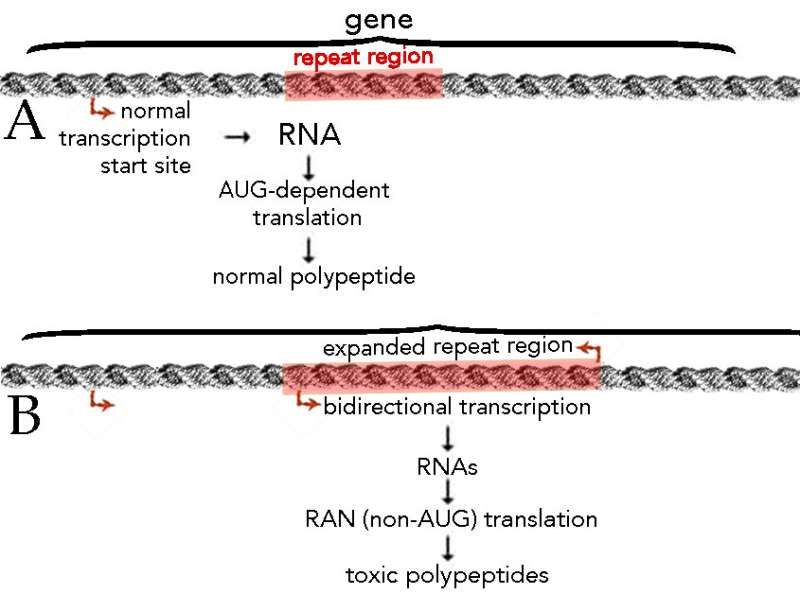
The situation is further complicated when the established rules of using RNAs to direct polypeptide synthesis via the process of translation, are violated, as occurs in what is known as "repeat-associated non-ATG (RAN)" polypeptide synthesis (see Cleary and Ranum, 2017). In this situation, the normal signal for the start of RNA-directed polypeptide synthesis, an AUG codon, is subverted – other RNA synthesis start sites are used leading to underlying or imbedded gene expression. This process has been found associated with a class of human genetic diseases, such as amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) characterized by the expansion of simple (repeated) DNA sequences (see Pattamatta et al., 2018). Once they exceed a certain length, such"repeat" regions have been found to be associated with the (apparently) inappropriate transcription of RNA in both directions, that is using both DNA strands as templates (← A: normal situation, B: upon expansion of the repeat domain). These abnormal repeat region RNAs are translated via the RAN process to generate six different types of toxic polypeptides.
So what are the molecular factors that control the various types of altORF transcription and translation? In the case of ALS and FTD, it appears that other genes, and the polypeptides and proteins they encode, are involved in regulating the expression of repeat associated RNAs (Kramer et al., 2016)(Cheng et al., 2018). Similar or distinct mechanisms may be involved in other neurodegenerative diseases (Cavallieri et al., 2017).
So how should all of these molecular details (and it is likely that there are more to be discovered) influence how genes are presented to students? I would argue that DNA should be presented as a substrate upon which various molecular mechanisms occur; these include transcription in its various forms (directed and noisy), as well as DNA synthesis, modification, and repair mechanisms occur. Genes are not static objects, but key parts of dynamic systems. This may be one reason that classical genetics, that is genes presented within a simple Mendelian (gene to trait) framework, should be moved deeper into the curriculum, where students have the background in molecular mechanisms needed to appreciate its complexities, complexities that arise from the multiple molecular machines acting to access, modify, and use the information captured in DNA (through evolutionary processes), thereby placing the gene in a more realistic cellular perspective.
Provided by Public Library of Science
This story is republished courtesy of PLOS Blogs: blogs.plos.org.