July 23, 2018 feature
Cell Studio: Serious games for immunology
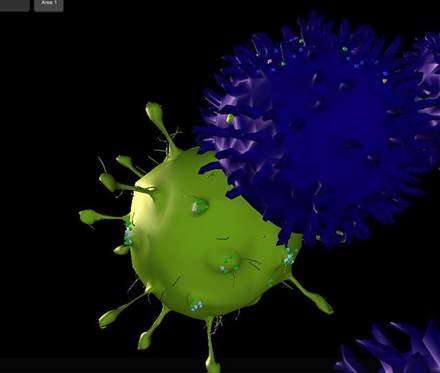
Researchers at the Tel Aviv University, Israel, Dortmund University, Germany, and Arizona State University, U.S., have collaborated to engineer an interactive 3-D simulation in silico that can mimic in vitro, or in vivo experiments as seen under the microscope using interactive game engines. The novel platform, known as Cell Studio, can simulate an organic microenvironment with biological and biophysical rules at the cellular level. Several biological scenarios can be simulated, including a 2-D or 3-D spatial patch of tissue or cell culture, cell mitosis, cell differentiation and apoptosis. This work was recently published in AIP Bioengineering.
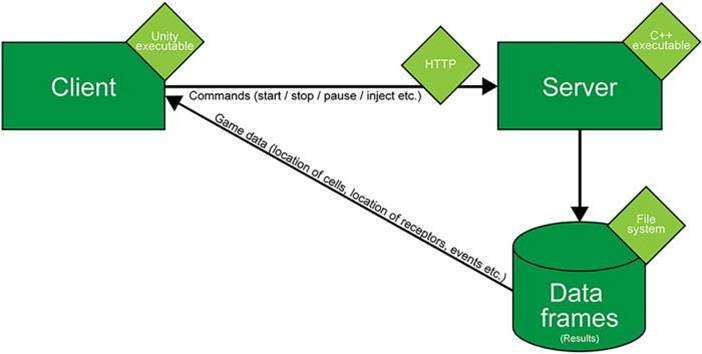
While the platform is aimed at biologists, biophysicists and researchers in computational biology and systems biology, Cell Studio does not require prior-training in coding; facilitating intuitive general use. The platform is available for download at www.cellstudio.info (currently compatible with Windows OS), initially developed by Liberman et al., to engineer a hybrid, cellular-level platform to model and simulate immunological processes with a user-friendly, graphical user interface (GUI). Researchers and educators can test a hypothesis prior to conducting a real experiment by simulating biological scenarios on the platform. Cell Studio is deployed via client-server architecture using the Unity 3-D (Unity Technologies) game engine at the client side and a scalable C++ algorithm on the side of the server. The use of game engines for work outside game development, with applications in architecture and medicine, is known as "serious games."
Users can feed biological data about an experimental system, run it in real-time and watch the simulation play out in 3-D, while interacting with it as intended. Rules of the simulation can be set by the user so that cells can respond to biological events, which include molecular secretions, changes to membrane protein expression, proliferation cues (mitosis) and apoptosis. Users can stop the experiment, change viewpoint, rewind or intervene by injecting molecules of any kind while the experiment runs. Additionally, while the simulation runs, numerical data becomes available in real-time via FACS plot displays, demonstrating receptor distribution on a monoclonal cell population and quantifying the number of cells in each monoclonal population as classic examples.
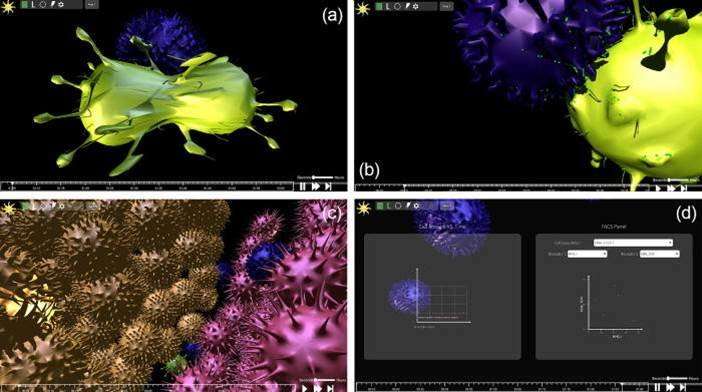
The front-end (user-interface) of the game-like simulation allows users to assign rules and visualize its run, built in to the Unity 3-D game engine using the Mega-Fiers Unity package. Cell Studio is based on a 'hybrid' biological modeling paradigm, which deconstructs a system to its constituent entities, assigning each cell or entity as an agent capable of making its own decision based on its local environment, while allowing user interaction with the simulation during the run.
Each cell model is engineered via a three-dimensional lattice (mesh) as its "skin" prior to the run. Compared to other such modeling programs, Cell Studio is notable, since visualized cell configurations occur in real time during the simulation according to user-defined constraints and conditions placed on the 3-D lattice of each cell type of interest. The experiments can also be run in batch mode and viewed offline. Furthermore, the simulation allows a "sanity check," whereby a modelled experiment can be validated against a real experiment to observe the accuracy of the modeling paradigm. The present version of Cell Studio can be implemented with cell templates in immunology, which includes antigen presenting cells (APCs), T cells and membrane receptors. Further upgrades are in progress to expand the biological niche and experimental scope of the simulation for scientists and educators.
The academic project is currently offering opportunities for game programmers, 3-D artists and research students interested in computational and systems biology to join the team and work to expand the domains of Cell Studio. Biophysicists and research biologists are encouraged to use the system and offer insights to upgrade the simulation. The project also seeks sponsors for funding. The project is in development under the guidance of the team comprising Dr. Uri Nevo (TAU), Dr. Sol Efroni and Prof. Ken Buetow. The project is written by Dr. Danny Kario, Ph.D. student Asaf Liberman and a team of interdisciplinary scientists.
More information: Asaf Liberman et al. Cell studio: A platform for interactive, 3-D graphical simulation of immunological processes, APL Bioengineering (2018). DOI: 10.1063/1.5039473
Katarzyna A. Rejniak et al. Hybrid models of tumor growth, Wiley Interdisciplinary Reviews: Systems Biology and Medicine (2010). DOI: 10.1002/wsbm.102
Journal information: Cell
© 2018 Phys.org