Cracking the RNA-binding code of a cell fate regulator
Interactions between RNA and proteins impact all aspects of RNA metabolism. These interactions are mediated by diverse RNA binding domains (RBDs), of which many remain to be discovered. One example of a recently identified RBD is the NHL domain. This domain is present in so-called TRIM-NHL proteins, like the C. elegans LIN-41 or its human homolog LIN41/TRIM71, collectively referred to as LIN41.
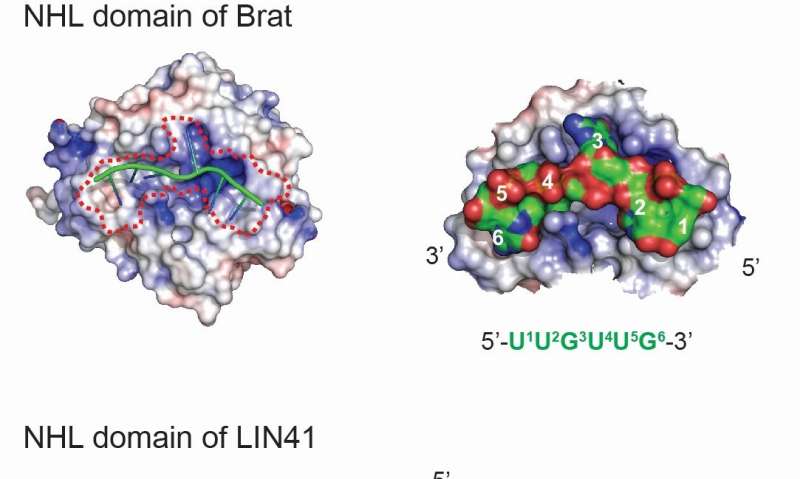
How exactly LIN41 proteins recognize specific RNAs remained unknown. Pooja Kumari, one of the authors leading this project, explains: "We knew that the NHL domain mediates the interaction of TRIM-NHL proteins with RNA: The NHL domain of Drosophila Brat binds RNA in a sequence specific manner and our previous work showed that also the NHL domain of LIN-41 mediates the RNA binding. However, for a long time we were unable to identify the RNA sequence motifs required for LIN41 binding."
A breakthrough came when Florian Aeschimann, another leading author, suggested that the binding could be mediated by secondary structures in the target mRNAs. Following up on this hypothesis, the FMI scientists, with computational input from Dimos Gaidatzis, could show that the NHL domain of LIN41 indeed interacts with particular RNA stem-loop (SL) elements, sharing only few sequence features but containing exactly three nucleotides in the loop.
Finally, with critical help from Jeremy Keusch and Heinz Gut from the protein structure facility, Kumari elucidated the crystal structure of LIN41-RNA complex: "From the crystal structure, we observed that the loop of the RNA SL is inserted into a positively charged cavity in the NHL domain and it's the RNA shape that defines the specificity of this interaction," said Kumari.
These findings came as a surprise as Kumari points out: "The identified form of interaction is entirely different from the RNA binding by Brat, otherwise a fairy similar protein. A plausible explanation for such RNA binding plasticity came from our collaborators from IIMCB in Warsaw, who, based on phylogenetic analyses, suggested that the different strategies to RNA binding reflect a rapid evolution of the NHL domain."
Rafal Ciosk said, "This work is a powerful example of how the combined use of biochemical, structural, computational and functional approaches can uncover the biological roles and evolutionary history of novel RNA-binding folds, paving a road to more such discoveries in the future."
More information: Pooja Kumari et al. Evolutionary plasticity of the NHL domain underlies distinct solutions to RNA recognition, Nature Communications (2018). DOI: 10.1038/s41467-018-03920-7
Journal information: Nature Communications