Computer models combat malaria

As one of the world's deadliest pathogens, Plasmodium spreads relentlessly from host to host. But a computer model created by KAUST scientists may reveal and help exploit the parasite's unknown weaknesses to uncover new options for treating malaria.
The Plasmodium parasite needs both a mosquito and vertebrate host to reproduce and complete its life cycle. Through what would otherwise be harmless insect bites, Plasmodium rapidly multiplies and spreads from mosquitoes to human hosts, where it causes malaria and kills nearly half a million people worldwide each year.
As Plasmodium infects red blood cells, transmits between hosts and reproduces, the single-celled parasite takes different forms. A key limitation of current malaria drugs is that they only treat the form of Plasmodium that actively causes malaria symptoms. Yet, treatments that target the pathogen's transmitting form may nip the disease in the bud.
Searching for potential malaria drug targets, KAUST Ph.D. student Alyaa Abdel-Haleem and her colleagues investigated the Plasmodium genetic blueprint. They focused on the genes involved in metabolism—the chemical reactions that allow the parasite to grow, reproduce and respond to its environment.
"Our aim in this study was to provide a model that catalogues metabolic differences between species and life-cycle stages to explore novel treatment strategies and targets," explains Abdel-Haleem.
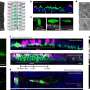
First, the scientists created computer models that combined the genetic blueprint of Plasmodium with information on how metabolic genes turned on and off in five different species and at five different stages of its life cycle. They then deleted genes in their models and ran simulations to test which genes were integral to the parasite's survival.
The scientists have already come across several potential targets. For one, the simulations revealed that in its symptom-causing form, the B vitamin pantothenate is an essential vitamin the Plasmodium falciparum species require for replication, while the same factor is not crucial for the growth of the malaria species that infect rodents.
"A potential strategy is for vaccination to block this important pathway for the parasite's growth and to limit its survival and propagation," Abdel-Haleem says.
Abdel-Haleem and her supervisors, Takashi Gojobori and Xin Gao, with computational support from Katsuhiko Mineta hope their Plasmodium simulation model will help uncover even more drug treatment leads. They highlight that the models can be used to translate findings across experimental models and human-infecting species.
Meanwhile, the team will continue to add more genetic information about Plasmodium as it becomes available. "The most crucial step is to keep the model updated for future use," says Gojobori.
More information: Alyaa M. Abdel-Haleem et al. Functional interrogation of Plasmodium genus metabolism identifies species- and stage-specific differences in nutrient essentiality and drug targeting, PLOS Computational Biology (2018). DOI: 10.1371/journal.pcbi.1005895
Journal information: PLoS Computational Biology