Solving the structure of ATP synthase
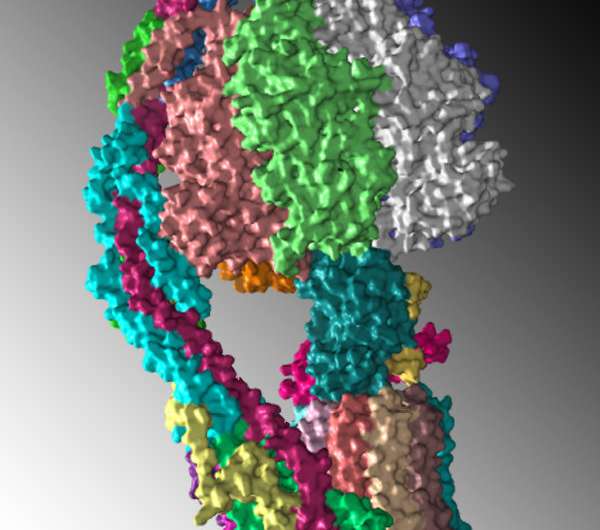
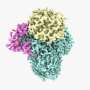
A team of scientists headed by Rosalind Franklin University Professor David M. Mueller, PhD, has solved the structure of mitochondrial ATP synthase, an enzyme that makes ATP, adenosine triphosphate, the major energy source of cells.
Given the importance of ATP synthase in fueling the cell, determining the complete structure of the enzyme has been a scientific holy grail, promising to reveal secrets about the ATP machine that could lead to therapeutic target identification and advance understanding of metabolic disease and other cellular pathologies.
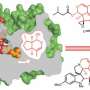
"Understanding how the enzyme actually works requires the knowledge of its three dimensional molecular structure at the atomic level," said Dr. Mueller, principal investigator for the study that used cryo-electron microscopy (cryo-EM) to reveal the enzyme at near atomic resolution.
The first complete structure of ATP synthase provides evidence for the mechanism by which the drug oligomycin inhibits the enzyme and how disease-causing mutations disrupt the function of the molecule. Solving the structure overcomes a barrier to understanding its likely broader function in disease and drug mechanisms.
The study, published by the journal Science via First Release on April 12, paves the way for determining the structures of other functional states of the ATP synthase, which has been the subject of structural studies for decades in labs across the globe.
Dr. Mueller, an investigator in the university's Center for Genetic Disease, took a different approach in collaboration with scientists from the National Institutes of Health and a team at Harvard University. They used cryo-EM analysis to decipher the engineered ATP synthase, which was synthesized in yeast and reconstituted into nanodiscs to allow for structural analysis. While cryo-EM isn't new, advancements in technology have made it possible to solve the structure at near atomic resolution.
"This study illustrates how a coordinated effort by scientists can bring about an important goal," Dr. Mueller said. "None of us were looking at this project from our own particular point of view. We were focused on the goal itself. Through collaboration we solved a problem that individually, we couldn't."
More information: Anurag P. Srivastava et al, High-resolution cryo-EM analysis of the yeast ATP synthase in a lipid membrane, Science (2018). DOI: 10.1126/science.aas9699
Journal information: Science