New active ingredients from the toolbox
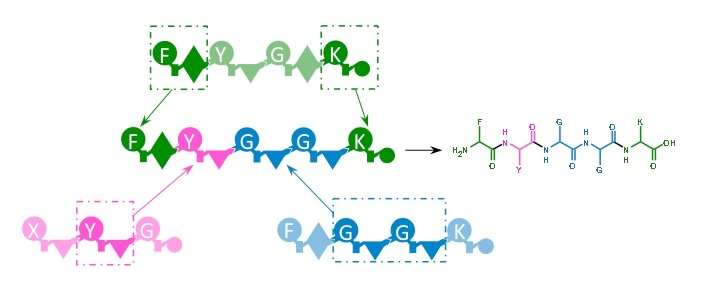
Microorganisms often produce natural products in a step-by-step manner similar to an assembly line. Examples of such enzymes are non-ribosomal peptide synthetases (NRPS). Researchers at Goethe University Frankfurt have now succeeded in designing these enzymes in such a way that they can produce completely new natural products.
Many important therapeutics, such as antibiotics or immunosuppressant and anti-cancer drugs, are derived from microorganisms. This is also the case for several different peptides which are produced in the microbial cell with the help of the NRPS enzymes. An NRPS functions like an assembly line in a modern car factory: new parts are added to the basic chassis at each workstation until a finished car rolls out of the plant at the end. In the case of the NRPS, a certain amino acid is selected, activated and processed at each station (known as modules) so that linear, cyclic or further modified peptides emerge at the end that can also carry unusual amino acids.
Although the fundamental principles of NRPS are long known, to date it was hardly possible to modify these enzymes. In the few cases where single modules were successfully exchanged, production of the modified natural products noticeably decreased. Assembling completely new enzymes, which in turn would produce completely new natural products, seemed totally impossible. The research group led by Professor Helge Bode, Merck Endowment Professor for Molecular Biotechnology at Goethe University Frankfurt, has now achieved this.
"In principle, we use natural NRPS systems from bacteria only as building blocks which we reassemble in a new way using new interfaces we've identified," says Bode, explaining the research approach. Yields are comparable with the natural production of these natural substances.
The method is meanwhile so sophisticated that even beginners can use it to produce new peptides and thus potential drugs shortly after a basic introduction. However, it has been a long road. "I was lucky that I had a team working with me on this project that did not allow itself to become discouraged, was very diligent and able to think outside the box," says Bode. "The interface we finally selected to assemble the individual building blocks is such that the classical modular order of the biosynthesis is no longer respected."
The next step is to modify first clinical drugs with this method and use biotechnology to produce them. Moreover, new information regarding the structure of these NRPS will be gathered as part of the LOEWE research focus MegaSyn led by Bode and Professor Martin Grininger, also from Goethe University Frankfurt. This will make it possible to further improve the method in order to allow the modification of related classes of natural products or even to produce whole libraries of natural products. First results are very promising.
More information: Kenan A. J. Bozhüyük et al. De novo design and engineering of non-ribosomal peptide synthetases, Nature Chemistry (2017). DOI: 10.1038/nchem.2890
Journal information: Nature Chemistry
Provided by Goethe University Frankfurt am Main