3-D packaging of DNA regulates cell identity
The fundamental mechanisms governing how cells form an identity such as becoming a muscle cell or a nerve cell are not fully understood. Multiple diseases, including cancer, have been linked to cells going down the wrong developmental path during maturation. A new study from the Perelman School of Medicine at the University of Pennsylvania suggests that the ability of a stem cell to differentiate into cardiac muscle (and by extension other cell types) depends on what portions of the genome are available for activation, which is controlled by the location of DNA in a cell's nucleus.
"The basis of this study is understanding the ability of a cell to respond to molecular cues to correctly become one cell type or another," said senior author, Rajan Jain, MD, an assistant professor of Cardiovascular Medicine. "We wanted to know how that is achieved, step by step, because stem cells, capable of becoming any cell type in the body, give rise to cardiac muscle cells." Jain and co-senior author Jonathan A. Epstein, MD, the executive vice dean and chief scientific officer at Penn Medicine, published their findings this week in Cell.
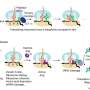
The study also suggests that knowing how to control how quickly a cell differentiates as it matures has important implications for regenerative medicine.Some regions of the genome are unavailable to be expressed because they are packaged tightly against the inner membrane of the cell nucleus (the lamina). These sequestered and silenced regions of DNA are called Lamin Associated Domains, or LADs. The Cell study suggests that the specific regions of silenced DNA at the periphery help define a cell's identity. For example, if nerve cell genes are held silent as LADs they cannot be expressed, so the cell does not become a neuron. However, if heart cell genes are released and available to be expressed, as happens during heart development, then those cells become cardiac muscle. Cell biologists have known for many years that some DNA is found near the inner nuclear membrane, but the function of this localization has been unclear. "Our work suggests that a cell defines its identity by storing away in an inaccessible closet the critical genes and programs necessary for it to mature into another cell type," Jain said. "In other words, a cell is 'who' it is because it has silenced 'who' it isn't." The Penn team found that an epigenetic enzyme called histone deacetylase (Hdac3) tethers DNA to the nuclear periphery. "We asked: Does this choreographed control of DNA availability contribute to a cell becoming a certain type?" Jain said. When they removed Hdac3 in stem cells during heart cell differentiation, they untethered regions of DNA containing heart-specific genes, allowing those genes to be activated, which led to precocious, too-fast differentiation.
"The implications of this study are far-reaching," Epstein said. "The ability to control how quickly a cell differentiates to make cardiac tissue or other cell types has important implications for regenerative medicine." In addition, in many diseases, including cancer, cells express genes that they normally would not, which changes their identity.
The study also addresses a classic concept in stem cell and developmental biology called "competence" - the ability of a cell to respond to its environment in specific ways. For example, some lung cells respond to cigarette smoke to become cancerous, while others do not. The investigators surmise that this difference could be due to the availability of regions of the genome to respond to chemicals associated with cigarette smoke, or because the unavailability of those same genes in non-responding cells are locked away in silenced domains at the nuclear periphery.
Jain, Epstein, and others are working to determine if changes in genome domains at the nuclear periphery, or the molecular tethers that keep them there, are responsible for cancer susceptibility. This approach could also be applied to other diseases, such as several forms of muscular dystrophy, heart failure, and premature aging due to inherited, genetic abnormalities of the lamina. "We aim to determine if these mutations lead to abnormal tethering of DNA and changes in gene expression and disease."
In the future, the researchers plan to manipulate the spatial organization of DNA to coax cells to adopt a different identity and ask what role that may play in human diseases linked to a loss of cellular identity, including diabetes, Alzheimer's disease, forms of heart failure, and cancer. The group is also expanding their work to study patients with mutations in components of their nuclear lamina.
Journal information: Cell