Microscope invented at marine biological laboratory illuminates chromosomal 'dark matter'
Using a microscope invented at the Marine Biological Laboratory (MBL), a collaborative team of biologists, instrument developers, and computational scientists has for the first time measured the density of a relatively inscrutable, highly condensed form of chromosomal material that appears in the cells of human beings and other eukaryotes. MBL scientists Michael Shribak (the microscope's inventor) and Tomomi Tani, together with Kazuhiro Maeshima of the National Institute of Genetics, Japan, recently reported their findings in Molecular Biology of the Cell.
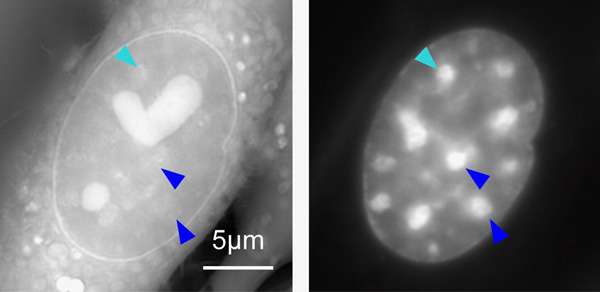
The scientists measured the density of heterochromatin, a tightly packed form of chromatin that appears as dark, scattered regions in the cell nucleus. Until recently, this chromosomal "dark matter" was thought to contain either noncoding DNA or silenced genes; however, new research suggests that heterochromatin DNA is not, in fact, fully inactive. To investigate this possibility, the physical properties of heterochromatin need to be described in live cells, which has been a significant challenge using traditional microscopy. The team was able to measure the density of heterochromatin in its natural state using a novel type of microscopy, orientation-independent differential interference contrast (OI-DIC), which Shribak first developed in collaboration with MBL Distinguished Scientist Shinya Inoué in the mid-2000s and has continued to improve.
This study, Shribak said, is "the first important application of OI-DIC," a technology that "is ideal for studying structure and motion in unstained, living cells and isolated organelles, because they can be followed for long periods of time non-invasively."
"This research exemplifies the kind of successful and productive interaction between biologists, microscope developers, and data scientists that is a central feature of MBL science, and is a major area of growth at the MBL," said David Mark Welch, director of the Marine Biological Laboratory Division of Research.
Widely used by biologists since the 1970s, conventional DIC microscopy uses beam-shearing interference to generate contrast-based images of live, unmodified cells and tissues. In the early 1980s at MBL, Shinya Inoué and Robert and Nina Allen of Dartmouth College independently invented video-enhanced DIC, which drastically improved the technique and its resolution. However, DIC suffered still from the drawback that the scientist had to rotate the biological sample several times to get a complete image, because cellular structures along the beam-shearing plane are invisible. In 2002, Shribak proposed orientation-independent DIC (U.S. patents 7233434 and 7564618), which solved the problem. The OI-DIC microscope rapidly takes images in different beam-shear directions, then processes a final image. It has the added advantage over conventional DIC of allowing quantitative measurement of the sample.
Last summer, Thomas Rhines, a University of Chicago student studying with Shribak as a Jeff Metcalf Undergradate Scholar, developed a method to measure the resolution of optical microscopes, focusing particularly on the OI-DIC. "This microscope provides the best possible resolution and contrast [of DIC microscopes]," Shribak said (about 250 nm at highest resolution). Rhines and Shribak will continue to collaborate to develop the best algorithm for data analysis of resolution. Shribak is also collaborating with MBL Fellow Patrick La Rivière of the University of Chicago to develop a 3D orientation-independent DIC system.
More information: Ryosuke Imai et al, Density imaging of heterochromatin in live cells using orientation-independent-DIC microscopy, Molecular Biology of the Cell (2017). DOI: 10.1091/mbc.E17-06-0359
Journal information: Molecular Biology of the Cell
Provided by Marine Biological Laboratory