New ultrafast method for determining antibiotic resistance
Researchers at Uppsala University have developed a new method for very rapidly determining whether infection-causing bacteria are resistant or susceptible to antibiotics. The findings have now been published in the U.S. journal Proceedings of the National Academy of Sciences (PNAS).
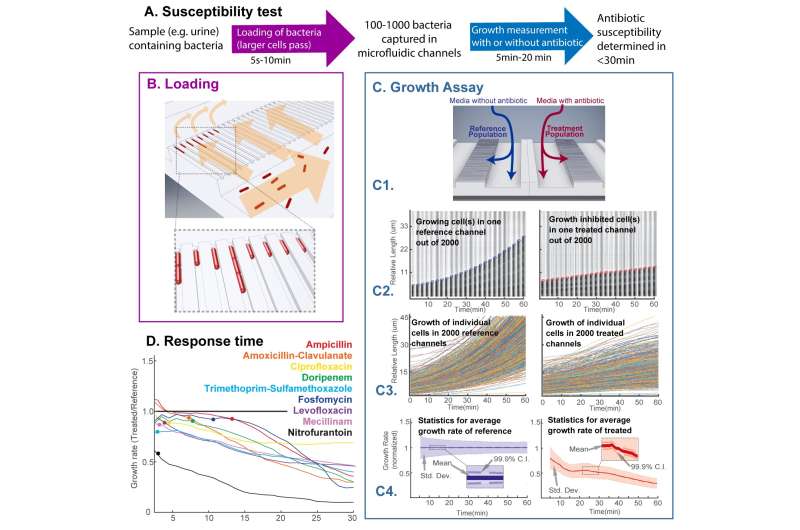
Antibiotic resistance is a growing medical problem that threatens human health globally. One important contributory factor in the development of resistance is the incorrect use of antibiotics for treatment. Researchers therefore seek reliable methods to quickly and easily identify bacterial resistance patterns, known as antibiotic susceptibility testing (AST), and provide early treatment, i.e. right from the doctor's appointment. This has been inhibited by the current time-consuming antibiotic resistance tests. Now, researchers at Uppsala have, for the first time, developed an antibiotic resistance test that is fast enough to inform antibiotic choice in the doctor's office. The test is primarily intended for urinary tract infections—a condition that, globally, affects approximately 100 million women a year and accounts for 25 per cent of antibiotic use in Sweden.
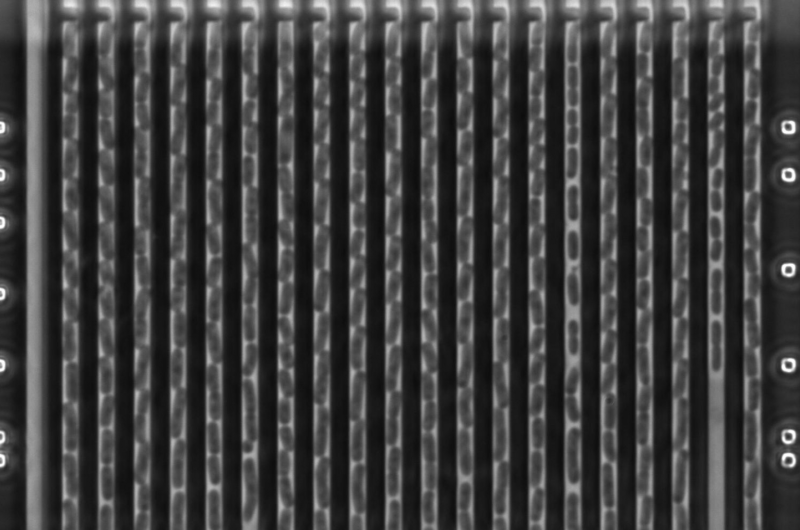
"We've developed a new method that allows determination of bacterial resistance patterns in urinary tract infections in 10 to 30 minutes. By comparison, the resistance determination currently in use requires one to two days. The rapid test is based on a new plastic microfluidic chip where the bacteria are trapped and methods for analysing bacterial growth at single-cell level," says PhD student Özden Baltekin, who performed most of the experimental work.
The "fASTest" method is based on sensitive optical and analytical techniques developed to study the behaviour of individual bacteria. Monitoring whether individual bacteria grow in the presence of antibiotics (i.e. are resistant) reveals their resistance or susceptibility within a few minutes.
"It's great that the research methods we developed to address fundamental questions in molecular biology are useful for such a tremendously important medical application," says Johan Elf, one of the researchers behind the study.
The detection method is now being developed by an Uppsala company, Astrego Diagnostics AB, into a user-friendly product. The company expects to have an automated test for urinary tract infections within a few years.
"The hope is that, in future, the method could be used in hospitals and health centres to quickly provide correct treatment and reduce unnecessary use of antibiotics," says Dan Andersson, one of the researchers behind the study.
"We believe the method is usable for other types of infection, such as blood infections where prompt, correct choice of antibiotic is critical to the patient."
More information: Özden Baltekina, et al., Antibiotic susceptibility testing in less than 30 min using direct single-cell imaging, PNAS, DOI: 10.1073/pnas.1708558114 , www.pnas.org/content/early/201 … 8/07/1708558114.full
Journal information: Proceedings of the National Academy of Sciences
Provided by Uppsala University