Bacterial classification may be more elusive than previously thought
New research from Dartmouth College raises questions over how scientists should interpret observed groupings of bacteria. The study advises caution with the assumption that bacterial clusters are always a result of ecological and genetic forces.
The research, appearing in the Proceedings of the National Academy of Sciences, says random diversification and extinction of cells could organize bacteria into taxonomic units just as effectively as classification based on selection-driven ecological forces.
"A reliable classification system is the key to understanding microbial biodiversity," said Olga Zhaxybayeva, assistant professor of biological sciences at Dartmouth College. "Through our research, we found that organizing microorganisms is even trickier than previously thought."
Scientists are currently divided over what factors to consider when classifying bacteria and other microorganisms. Some favor the so-called "periodic selection" model, in which the descendant of the most-fit genotype takes over the population and establishes a new group. Others advocate the "recombination" model, in which the frequent exchange of material between genes within bacterial populations causes organisms to cluster.
"Not knowing what is driving the organization of microorganisms makes the task of providing fast, accurate identification of bacteria difficult," said Zhaxybayeva. "Surprisingly, we found that a simple alternative may also explain grouping patterns, eliminating the need to invoke current models."
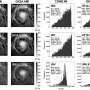
The research team, led by Zhaxybayeva, tested the idea that a simple birth-death cycle of cells can produce microbial clusters that look like groupings observed in nature. Analysis of hundreds of genomes within four bacterial groups - Escherichia spp., Borrelia spp., Neisseria spp. and Helicobacter pylori - produced patterns indistinguishable from those observed in most genes from three of the four bacterial groups.
As a result of the findings, the study recommends checking diversification of microbial groups against the proposed birth-death model before calling for more complex explanations.
More information: Timothy J. Straub el al., "A null model for microbial diversification," PNAS (2017). www.pnas.org/cgi/doi/10.1073/pnas.1619993114
Journal information: Proceedings of the National Academy of Sciences
Provided by Dartmouth College