Study targets LigM for its role in breaking down aromatic pollutants
A protein used by common soil bacteria is providing new clues in the effort to convert aryl compounds, a common waste product from industrial and agricultural practices, into something of value.
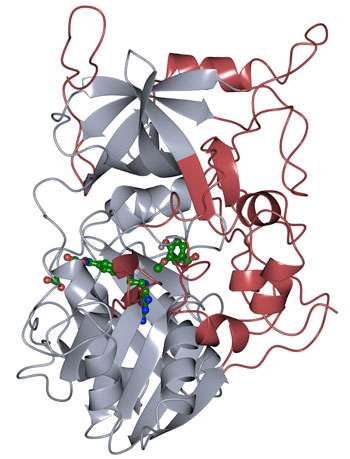
Researchers at the Department of Energy's Lawrence Berkeley National Laboratory (Berkeley Lab) and Sandia National Laboratories working at the Joint BioEnergy Institute (JBEI) have resolved the protein structure of the enzyme LigM, which is utilized by the soil bacterium Sphingomonas to metabolize aryl compounds derived from lignin, the stiff, organic material that gives plants their structure.
Their work is reported today in the Proceedings of the National Academy of Sciences.
In biofuel production, aryl compounds are a byproduct of the breakdown of lignin. Many of the pathways leading to the breakdown of lignin involve demethylation, which is often a critical precursor to any additional steps in modifying lignin-derived aryl compounds.
Study lead author Amanda Kohler, JBEI postdoctoral researcher at Sandia, noted that LigM is an attractive demethylase for use in aromatic conversion because it is a simple, single-enzyme system. LigM is also able to maintain its functionality over a broad temperature range.
"When we're trying to build new pathways in synthetic biology, the simpler the system the better," said Kohler.

The researchers found that half of the LigM enzyme was homologous to known structures with a tetrahydrofolate-binding domain that is found in simple and complex organisms alike. The other half of LigM's structure is completely unique, providing a starting point for determining where its aryl substrate-binding site is located. They also figured out that LigM is a tyrosine-dependent demethylase.
"It's the first of its kind to be identified," said Kohler. "This research provides the much-needed groundwork to help in the development of an enzyme-based system for converting aromatic waste products into something useful."
Kohler said they are now working on engineering LigM so that it is able to act on a wider range of aryl substrates in addition to targeting specific aryl waste products.
More information: Amanda C. Kohler et al. Structure of aryl-demethylase offers molecular insight into a catalytic tyrosine-dependent mechanism, Proceedings of the National Academy of Sciences (2017). DOI: 10.1073/pnas.1619263114
Journal information: Proceedings of the National Academy of Sciences
Provided by Lawrence Berkeley National Laboratory