Mechanism for hMTH1's broad substrate specificity revealed
Human MutT homolog 1 (hMTH1) protein acts as the primary enzyme for breaking down (hydrolyzing) damaged (oxidized) deoxynucleoside triphosphates (dNTPs) the substrates of DNA synthesis. Recently, hMTH1 has drawn attention as a popular target for new anticancer therapies because it is non-essential for normal cells, but cancer cells require it to avoid incorporating oxidized nucleotides into DNA, which would result in cancer cell death. Now, hMTH1 inhibitors are developed as anticancer drug candidates. Even though some reports argue against the usefulness of hMTH1 inhibition, highly potent and selective inhibitors of hMTH1, which would allow for the introduction of oxidized nucleotides into cancer cell DNA, are expected for future cancer treatment.
Enzymes are usually very particular about the material they catalyze (substrate). Some, however, can catalyze more than one substrate, and hMTH1 is known to hydrolyze several oxidized dNTPs. The reasons behind the broad substrate specificity had not yet been explored when researchers from Japan began considering the issue.
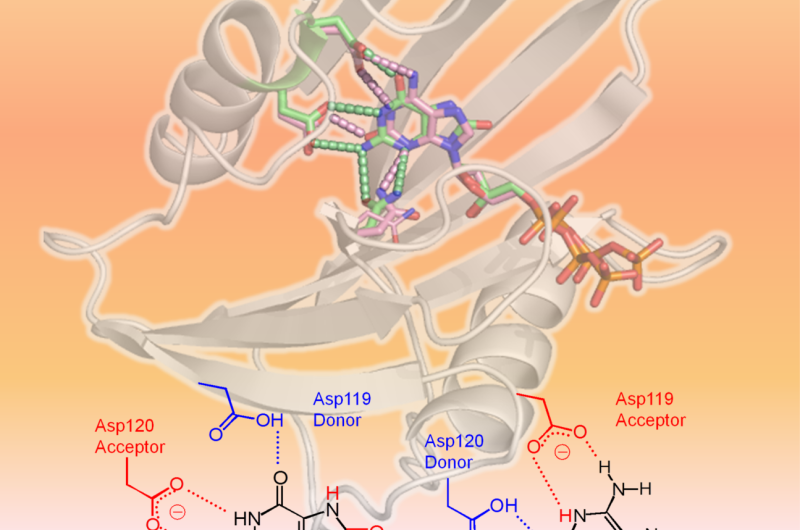
Scientists from Kumamoto University, the National Institutes for Quantum and Radiological Science and Technology, and Kyushu University performed structural and kinetic analyses on hMTH1 to determine the mechanisms behind its ability to hydrolyze various oxidized mutagenic dNTPs, such as 8-oxo-dGTP and 2-oxo-dATP, with similar efficiency. Their experiments found evidence that the protonation state between the aspartate residues Asp-119 and Asp-120 was the determining factor in hMTH1's broad substrate specificity. In other words, the addition or removal of one or more protons at Asp-119 or Asp-120 is what causes hMTH1 to recognize 8-oxo-dGTP or 2-oxo-dATP. This allows hMTH1 to hydrolyze the oxidized mutagenic dNTPs before they are used for DNA synthesis; DNA that contains oxidized nucleotides would cause cancer cell death.
"We found that differing protonation states between Asp-119 and Asp-120 is the signal for hMTH1 to hydrolyze the damaged nucleotides. If this action by hMTH1 can be suppressed, cancer cells would accumulate damaged nucleotides and eventually undergo apoptosis," said Kumamoto University's Professor Yuriko Yamagata, leader of the research group. "The clarification of this mechanism should help in the development of hMTH1 targeting anticancer drugs."
More information: Shaimaa Waz et al, Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity, Journal of Biological Chemistry (2017). DOI: 10.1074/jbc.M116.749713
Journal information: Journal of Biological Chemistry
Provided by Kumamoto University