For biologists studying tiny worms, new technologies make big improvements
For decades, the tiny roundworm C. elegans has been a vital tool in the biomedical researcher's toolkit, proving central to groundbreaking discoveries such as green fluorescent protein, the molecular marker used universally across research labs. Now, scientists in the laboratories of Shai Shaham and Eric D. Siggia at Rockefeller University are pushing the envelope even further on what C. elegans can teach us. They are developing technologies to study new aspects of how organs and nervous systems develop in these useful creatures.
"With any new technology, what's exciting is that we can't predict what we'll discover," says Shaham, Richard E. Salomon Family Professor and head of the Laboratory of Developmental Genetics. "Now that new technologies are available, we have the possibility of identifying new biology."
Freeze and say cheese
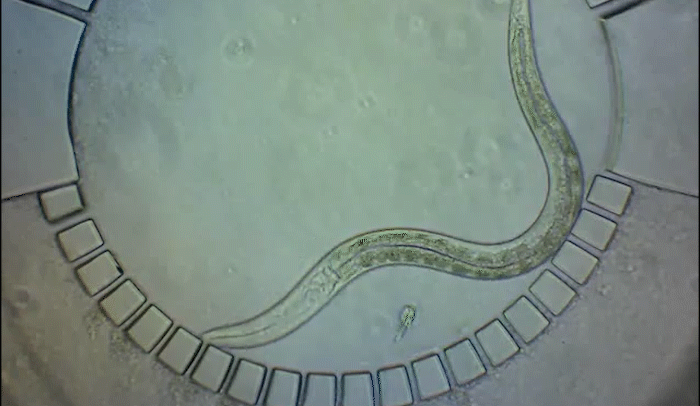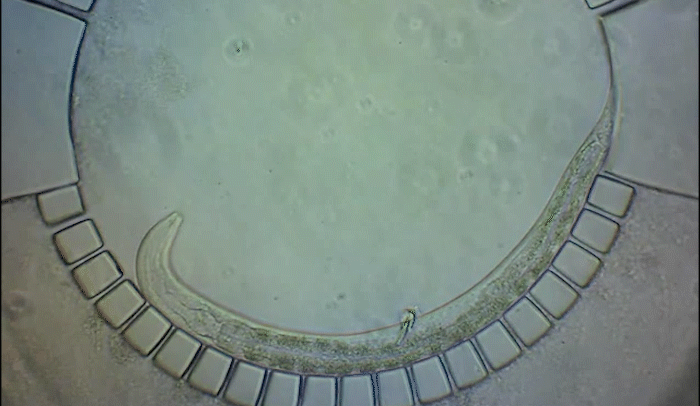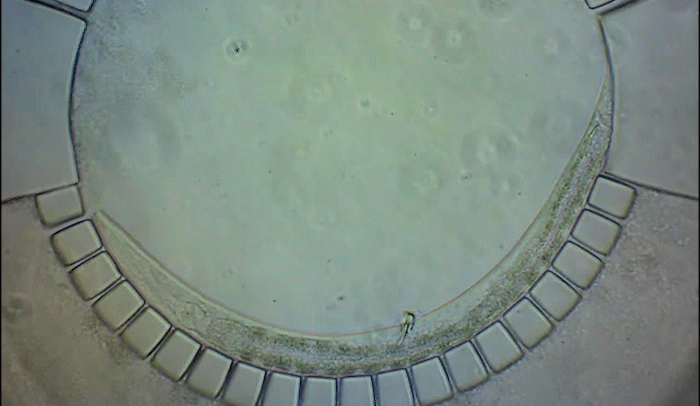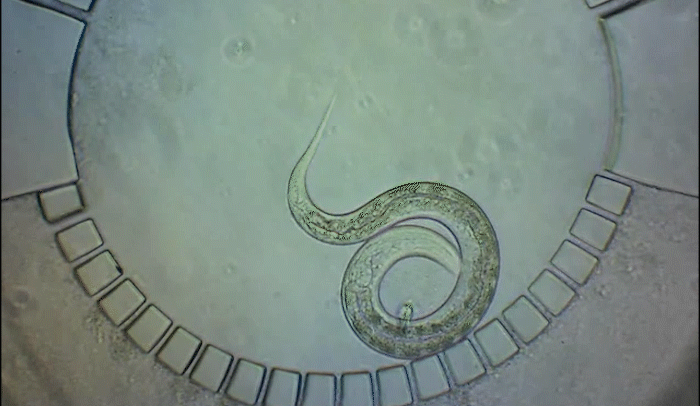
To pinpoint key steps in the worms' development, Wolfgang Keil, a postdoctoral fellow who shares his time between the labs of Shaham and Siggia, wanted to follow patterns of cell division in C. elegans larvae. But the existing methods were tedious and could only capture worms in an unnatural state—glued to a microscope slide. To solve this, Keil and colleagues invented a small chamber that allows the worm to move around and eat freely, except during short intervals when it is being imaged with a microscope.
The circular chamber is surrounded by posts that form tiny channels—just big enough to allow food to flow in, but too small for the worm to escape. When it is time to keep the worm still for imaging, a collapsible ceiling gently lowers over it, and the animal is simultaneously pushed to the side of the chamber for a short time—just enough to take a crisp snapshot. The technique is described in a study published in Developmental Cell.
"Embryos are isolated from the environment by an egg-shell, but that's not how subsequent larval development works," says Keil, lead author of the study. "What's most exciting about this method is that it enables you to go beyond what's been possible before—to study a developing animal that's feeding, growing, and possibly interacting with the environment."
Shaham says he and his colleagues are just at the tip of the iceberg in terms of what can be learned from this system, but they've already uncovered some surprises, including a rare variant in which a vital C. elegans organ is assembled a bit differently than usual. Siggia, Viola Ward Brinning and Elbert Calhoun Brinning Professor and head of the Laboratory of Theoretical Condensed Matter Physics, explains that observing this kind of natural variation was only possible because the data set collected was so large.
"This is a holy grail in the field," says Shaham, "because nobody has been able to create a system like this that's reliable." Using the new technology, 10 worms can be imaged at once, greatly improving upon the statistical power of earlier approaches. Previous methods, Shaham notes, "required us to sit in front of the microscope for hours and days, and putting the data together took years. Now we can essentially do the same thing in a single experiment."
The extended period over which they are able to image growing worms also allows the researchers to image long-term processes like cell death and the development of neurons, which they are just starting to explore.
Neurons take shape, one by one
Researchers in the Shaham laboratory have also developed innovative tools to study nervous system development more specifically, tracking one to four neurons at a time. The procedure involves turning on gene expression through a heat-sensitive protein that is activated when exposed to the heat of an infrared laser, and can be used to make fluorescent labels glow in specific neurons.
A recent study in Nature Communications describes the new method, the first to allow researchers to track the movement of any single neuron in a developing worm.
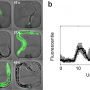
"With this system, you can label any cell or express any gene of interest, then use time-lapse imaging to track neurons as they grow," says Anupriya Singhal, a biomedical fellow and first author of the study. "It really opens the door to systematically characterizing how the nervous system develops at a resolution we've never been able to see before."
First, the researchers optimized the technology to use temperatures that didn't harm the worms. They found that exposing an embryonic worm cell to the laser for only five minutes activated the fluorescent label for up to six hours, which is enough to capture the major steps of neural development. They also identified a novel mechanism for neuronal growth and were able to confirm that a specific gene is vital for sensory organs to develop properly.
"Our goal is to eventually get a complete description of C. elegans neural development on a cell-by-cell basis, which may yield underlying principles that govern nervous system development elsewhere, including humans," says Shaham.
More information: Anupriya Singhal et al. Infrared laser-induced gene expression for tracking development and function of single C. elegans embryonic neurons, Nature Communications (2017). DOI: 10.1038/ncomms14100
Wolfgang Keil et al. Long-Term High-Resolution Imaging of Developing C. elegans Larvae with Microfluidics, Developmental Cell (2017). DOI: 10.1016/j.devcel.2016.11.022
Journal information: Nature Communications , Developmental Cell
Provided by Rockefeller University