'Smart' bacteria remodel their genes to infect our intestines
Infectious diarrhea, a common disease of children, is responsible for over 2 million infant deaths annually in developing counties alone. A primary cause of this and other devastating conditions is enteropathogenic bacteria, which attack the intestinal tract when contaminated food is consumed.
The infection process involves hundreds of genes and proteins, both in the infectious bacteria and the human host. However, the processes by which the pathogens establish themselves in our gut are poorly understood.
Now, a new study published in the prestigious journal Science, by researchers at the Hebrew University of Jerusalem's Faculty of Medicine, describes how pathogens sense their host, and tailor their gene expression to exploit their host to cause disease. The research was led by led by Prof. Ilan Rosenshine, the Etta Rosensohn Professor of Bacteriology at the Hebrew University.
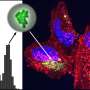
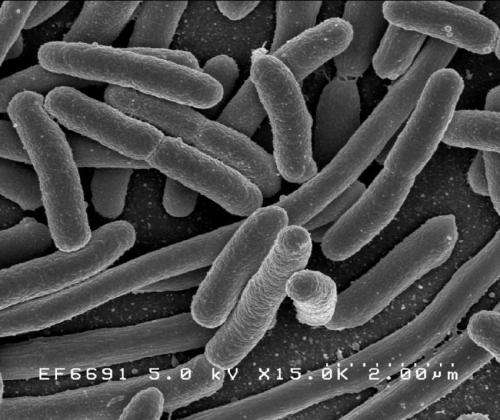
Working with a pathogenic strain of E. coli, the researchers found that the bacteria can sense attachment to the human intestinal cells and activate gene expression in response. This was demonstrated by engineering one of these genes to express a protein that stains the expressing bacteria to appear green under the microscope. Under microscopic examination, the researchers observed that only the attached bacteria fluoresce in bright green, whereas non-attached bacteria remain dark.
The researchers also deciphered how upon sensing that it has attached to intestinal cells, the pathogen reorganizes its gene expression, including genes involved in virulence and metabolism, to exploit the host cell. These findings may lead to the development of new strategies to combat bacterial infection.
"The next steps include mapping in detail the genes that change their expression upon attachment, and describing the precise effects of this expression remodeling," said Prof. Ilan Rosenshine. "Another important issue is testing whether similar regulation is involved in the infection processes of other pathogens."
More information: Naama Katsowich et al. Host cell attachment elicits posttranscriptional regulation in infecting enteropathogenic bacteria, Science (2017). DOI: 10.1126/science.aah4886
Journal information: Science
Provided by Hebrew University of Jerusalem