On the origin of life: Studying how the first biomolecule self-replicated
It's the ultimate chicken-or-egg conundrum: What was the "mother" molecule that led to the formation of life? And how did it replicate itself? One prominent school of thought proposes that RNA is the answer to the first question. Now, in ACS Central Science, researchers in this camp demonstrate RNA has more flexibility in how it recognizes itself than previously believed. The finding might change how we picture the first chemical steps towards replication and life.
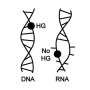
Today, plants, animals and other organisms reproduce by making copies of their DNA with the help of enzymes and then passing the copies onto the next generation. This is possible because genetic material is made of building blocks—or bases A, T, U, G and C—that pair up in a specific way. A pairs with T (or U in RNA), and G pairs with C. This rule is called Watson-Crick base pairing, named after the scientists who were credited with solving DNA's structure. But before life as we know it existed, some molecule had to replicate without any help at all.
RNA is a likely suspect for this go-it-alone first status because it is simultaneously capable of specific base-pairing like DNA, and catalyzing reactions, like an enzyme. Thus, Jack Szostak and colleagues wanted to investigate how RNA matches up with free nucleotides to see whether its base-pairing methods would allow RNA to copy itself without any outside aid.
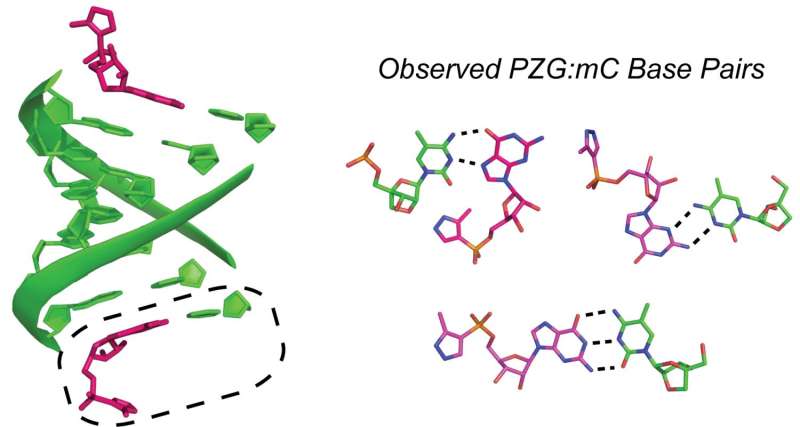
They monitored how an analogue of a free nucleotide interacted with a short piece of RNA using the classic method of X-ray crystallography—the same technique used more than fifty years ago in the original discovery of DNA's 3-D structure. In addition to forming the expected canonical Watson-Crick pairs, the RNA bonded with the analogue in other less frequently observed ways. Under prebiotic conditions, these unexpected non-Watson-Crick pairings might have caused dead-ends to replication. Thus, the results suggest that the first steps toward life required more trial and error than previously thought.
More information: Wen Zhang et al. Unusual Base-Pairing Interactions in Monomer–Template Complexes, ACS Central Science (2016). DOI: 10.1021/acscentsci.6b00278
Abstract
Many high-resolution crystal structures have contributed to our understanding of the reaction pathway for catalysis by DNA and RNA polymerases, but the structural basis of nonenzymatic template-directed RNA replication has not been studied in comparable detail. Here we present crystallographic studies of the binding of ribonucleotide monomers to RNA primer–template complexes, with the goal of improving our understanding of the mechanism of nonenzymatic RNA copying, and of catalysis by polymerases. To explore how activated ribonucleotides recognize and bind to RNA templates, we synthesized an unreactive phosphonate-linked pyrazole analogue of guanosine 5′-phosphoro-2-methylimidazolide (2-MeImpG), a highly activated nucleotide that has been used extensively to study nonenzymatic primer extension. We cocrystallized this analogue with structurally rigidified RNA primer–template complexes carrying single or multiple monomer binding sites, and obtained high-resolution X-ray structures of these complexes. In addition to Watson–Crick base pairing, we repeatedly observed noncanonical guanine:cytidine base pairs in our crystal structures. In most structures, the phosphate and leaving group moieties of the monomers were highly disordered, while in others the distance from O3′ of the primer to the phosphorus of the incoming monomer was too great to allow for reaction. We suggest that these effects significantly influence the rate and fidelity of nonenzymatic RNA replication, and that even primitive ribozyme polymerases could enhance RNA replication by enforcing Watson–Crick base pairing between monomers and primer–template complexes, and by bringing the reactive functional groups into closer proximity.
Journal information: ACS Central Science
Provided by American Chemical Society