Identifying brain regions automatically
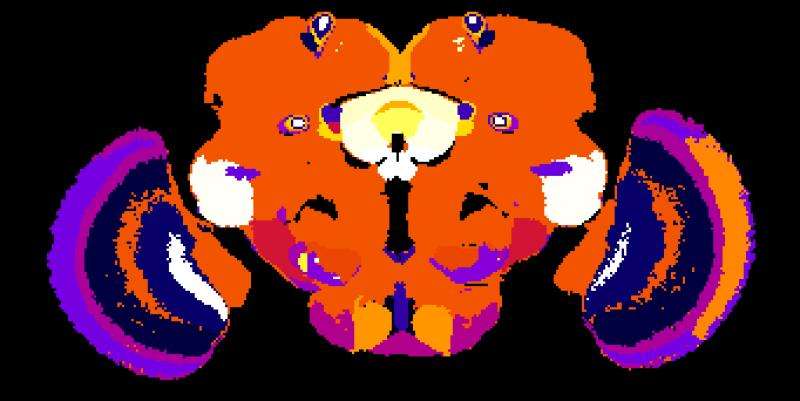
Using the example of the fruit fly, a team of biologists led by Prof. Dr. Andrew Straw has identified patterns in the genetic activity of brain cells and taken them as a basis for drawing conclusions about the structure of the brain. The research, published in Current Biology, was conducted at the University of Freiburg and at the Research Institute of Molecular Pathology (IMP) in Vienna, Austria.
The newly developed method focuses on enhancers, DNA segments responsible for enhancing transcription of RNA at specific locations and developmental times in an organism. The research started with a database of three-dimensional images showing individual enhancer activity. The team used an automatic pattern finding algorithm to identify genetic activity patterns shared across the images. They noticed that, in some cases, these patterns seemed to correspond with specific brain regions. To demonstrate the functionality of their method, the biologists began by applying it to regions of the fruit fly brain whose anatomy is already well known – namely, those responsible for the sense of smell. The activity patterns of the enhancers traced the already familiar anatomy of these regions.
Then the biologists used the new method to study brain regions responsible for vision. These experiments led to new insights into the anatomy of these areas: In addition to eleven already known regions, the activity patterns of the enhancers revealed 14 new regions, each of which presumably serves a different function for the fruit fly's sense of sight. The researchers now aim to conduct further studies to determine which regions are responsible for which functions.
Andrew Straw has served since January 2016 as professor of behavioral neurobiology and animal physiology at the University of Freiburg's Faculty of Biology and is a member of the Bernstein Center Freiburg (BCF). Before their move to Freiburg, he and his research assistants Karin Panser and Dr. Laszlo Tirian worked at the Research Institute of Molecular Pathology in Vienna in collaboration with Dr. Florian Schulze, Virtual Reality and Visualization Research Center GmbH (VRVis). The goal of Straw's research is to achieve a better understanding of the structure and function of the brain. He hopes this basic research will ultimately help in the design of therapies for patients suffering from neurological diseases affecting specific regions of the brain.
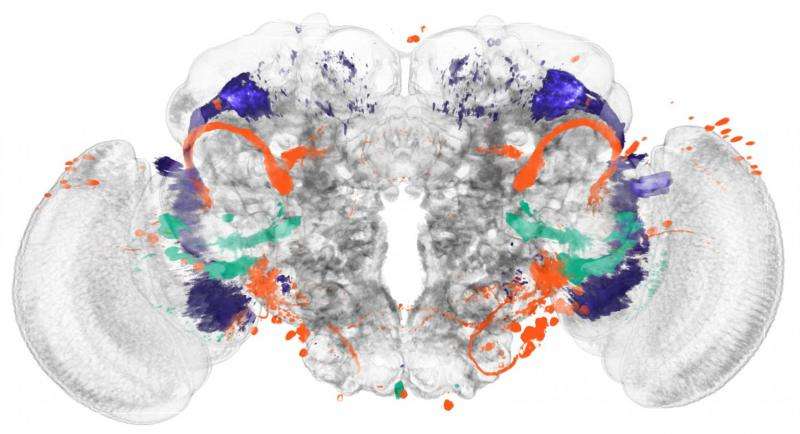
More information: Automatic segmentation of Drosophila neural compartments using GAL4 expression data reveals novel visual pathways. DOI: 10.1101/032292
Journal information: Current Biology
Provided by Research Institute of Molecular Pathology