Hormones in action: it's all about the right partner
Scientists at the Research Institute of Molecular Pathology (IMP) in Vienna uncover basic mechanisms of action for hormones.
Thousands of regulatory regions on the genomic DNA determine which part of a cell's genetic information is expressed and which is silent. Daria Shlyueva and Alexander Stark from the IMP in Vienna analysed such control-regions and the changes in activity that follow treatment with a hormone. They showed that - depending on the cell type - a single hormone can influence different regions. The findings are published in the advance online edition of the journal Molecular Cell this week.
The entire information for the development and functioning of an organism is encoded in its DNA. Genes contain the building plans for the molecules that constitute living beings. However, not all genes are active at all times, allowing the various cell types and organs to perform specific functions. This diversity is precisely regulated by non-coding regulatory regions in the genomic DNA. Alexander Stark, a biochemist and Group Leader at the IMP, is interested in finding out how these regions act as "molecular switches" to turn genes on or off at the right time.
STARR-Seq, reloaded
In 2013, the Stark group developed STARR-Seq, a novel technique to screen entire genomes for certain control regions of gene expression, so-called enhancers, and to measure their activity. Now they took this method one step further. "We were always interested in finding out what characterises a DNA-sequence that performs regulatory functions", he explains. "Now we wanted to go into more detail and show that STARR-Seq can identify hormone-dependent regulators and measure them."
Daria Shlyueva, a PhD student in the Stark group, cultivated two different cell types of the fruit fly and exposed them to the steroid hormone ecdysone. By comparing enhancer activities along the entire genomes of treated and untreated cells, the scientists were able to show that a single hormone can regulate different enhancers in different cells and can influence the activity of genes either positively or negatively.
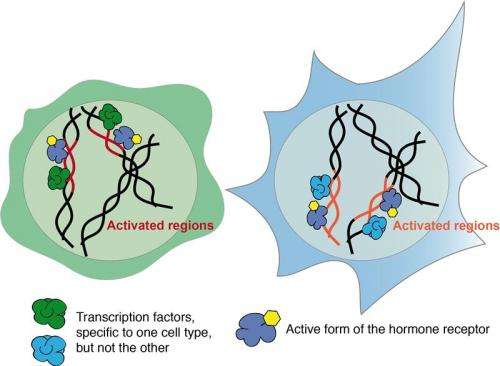
The hormone's receptor in the cell nucleus and further binding-partners, so-called transcription factors, determine which region of the DNA is influenced. The combination of these interacting molecules is responsible for the variable responses in different tissues and at different stages of development. These findings explain how regulatory regions of DNA can combine input from the cell's environment with cell type-specific information to trigger a whole range of reactions.
Searching for hormone-dependent DNA-elements in humans
Steroid hormones such as estrogen or testosterone have important functions in humans. Researchers in the lab of Alexander Stark have therefore started to look for molecular switches in the human genome. "In the future, our system could be used to identify control regions on DNA that are regulated by human hormones", says Stark. "This would be another important step towards potential medical applications."
More information: D. Shlyueva, C. Stelzer, D. Gerlach, J.O. Yáñez-Cuna, M. Rath, L.M. Boryń, C.D. Arnold and A. Stark: "Hormone-responsive enhancer activity maps reveal predictive motifs, indirect repression and targeting of closed chromatin." Mol. Cell, online Early Edition, 27 March 2014. DOI: 10.1016/j.molcel.2014.02.026
Journal information: Molecular Cell
Provided by Research Institute of Molecular Pathology