Keeping RNases poised for action
A better knowledge of RNA metabolism is key to understanding how RNAs regulate development and differentiation, and how their malfunction leads to disease. A team led by Helge Grosshans of the Friedrich Miescher Institute for Biomedical Research (FMI) has now identified a novel and evolutionarily conserved mechanism that preserves the stability of RNases and keeps them poised for RNA processing and degradation. The results have been published in Nature Structural & Molecular Biology.
For many decades, scientists thought that RNA functions merely as an intermediary between DNA and proteins. However, it has now become evident that RNA occurs in many flavors in the cell, allowing it to perform diverse and important functions. In addition to serving as a photocopy of DNA, a coupler between the genetic code and protein building blocks, and a structural component of ribosomes (the protein-producing machines), RNAs also directly regulate gene activity during development and differentiation. Accordingly, RNA malfunction has been implicated in numerous diseases, and research into RNA metabolism – the processes whereby RNA is formed, processed and degraded – has gained momentum.
RNases (ribonucleases) are key enzymes in RNA metabolism, as they can chew up RNAs partially or completely, thus causing either RNA maturation from precursors or inactivation by degradation. However, it has remained unclear how RNase activity is controlled. Helge Grosshans and his group at the FMI have now identified a mechanism – conserved between the roundworm C. elegans and higher vertebrates – that preserves the stability of RNases in the absence of substrate. In a paper published in Nature Structural & Molecular Biology, they describe how an RNase forms a complex with a protein partner, and how this complex formation prevents inactivation of the RNase.
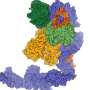
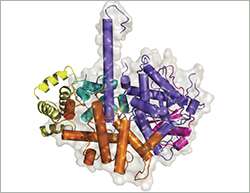
As a first step, Hannes Richter, a PhD student in the Grosshans laboratory, collaborated with a colleague from FMI's protein structure facility to elucidate the structure of a complex of the C. elegans RNase XRN2 and its partner PAXT-1. Richter, first author of the study, comments: "Our previous biochemical experiments already suggested that PAXT-1 interacted with XRN2 through its N-terminus. The crystal structure of XRN2 bound to PAXT-1 confirmed this: there are several amino acid residues in the N-terminus of PAXT-1 that mediate binding to XRN2. However, one amino acid in PAXT-1, Tyr56, is particularly important. When it is changed, the whole complex falls apart." In collaboration with a scientist from FMI's C. elegans facility, Richter showed that worms with a mutation in this tyrosine had the same symptoms as worms lacking PAXT-1 altogether.
Interestingly, the XRN2-binding domain (XTBD) also occurs in other vertebrate proteins which, apart from this domain, share little homology to PAXT-1. "The critical residue is highly conserved," says Richter. With this in mind, the scientists investigated whether three vertebrate proteins with XTBDs also interacted with XRN2 and could even functionally replace PAXT-1 in the worm. Richter reports: "The function of the XTB domain is indeed conserved: vertebrate XTBD-containing proteins bind to XRN2 in vitro, and a human XTBD protein rescued worms lacking PAXT-1."
But how does PAXT-1 binding to an RNase affect RNA metabolism? In a set of biochemical experiments, the scientists showed that PAXT-1 stabilizes XRN2 and preserves its activity in the absence of RNA. Grosshans notes: "This is something new – we tend to think of enzyme complexes as something that alters the activity of enzymes on their substrates. Here we find that PAXT-1 takes care of 'idle' XRN2 molecules, preventing them from going out of shape. This ensures that there is a constant pool of active RNases in the cell, poised for further action."
More information: Hannes Richter et al. Structural basis and function of XRN2 binding by XTB domains, Nature Structural & Molecular Biology (2016). DOI: 10.1038/nsmb.3155
Journal information: Nature Structural & Molecular Biology