Gone fishin' for natural products, with a new dragnet
Nature contains a treasure trove of substances that could help fight human disease. Just this year, the Nobel Prize in Physiology or Medicine honored the development of drugs that fight parasites and malaria based on such "natural products." But finding these molecules and discovering new chemical identities represents slow and painstaking work. This week in ACS Central Science, researchers report a new way to greatly speed up that process.
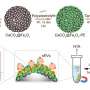
Current methods for sussing out new molecules are much like casting a fishing line in a vast ocean. And once a compound is on the hook, there's still much more work to be done to find out what it looks like and, ideally, how it is made. Because bacteria and other organisms make natural products using protein-based assembly lines, the genes that encode for them cluster together in the genome, making them easier to recognize. So, Neil Kelleher, William Metcalf and colleagues surmised that if they evaluated the amount of various natural products and compared that to the abundance of these assembly lines, they could determine which genes produced which natural products for many dozens of cases in parallel.
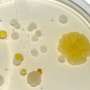
To demonstrate the new approach, the authors sequenced the genomes of 178 different strains of a bacterial family called the actinomycetes, while also measuring the amounts of the natural products made by each strain using mass spectrometry. When a natural product and a gene cluster involved in an assembly line were frequently found together in different species of bacteria, they scored this co-occurrence and inferred that they go together. This technique allowed the team to identify a new natural product they called tambromycin, along with its cellular machinery. Interestingly, tambromycin features a previously unknown amino acid they called tambroline, and the team showed it could slow the growth of two types of leukemia without affecting healthy cells. The authors note that their method improves with the number of strains interrogated and will standardize access to thousands of new lead compounds to reinvigorate drug discovery pipelines with natural molecules.
More information: "Metabologenomics: Correlation of Microbial Gene Clusters with Metabolites Drives Discovery of a Nonribosomal Peptide with an Unusual Amino Acid Monomer" pubs.acs.org/doi/full/10.1021/acscentsci.5b00331
Journal information: ACS Central Science
Provided by American Chemical Society